Proposal (633) to South American Classification Committee
Modify
linear sequence of genera and species in Emberizidae
Effect
on SACC: This would modify the linear sequence of
genera and species to reflect the recently published phylogeny of the family by
Klicka et al. (2014).
Background: The current sequence
is listed below. It is based largely on
historical momentum and reflects the transfer of many genera to the Thraupidae
and the transfer of Chlorospingus to
the family.
Melospiza lincolnii
Zonotrichia capensis
Ammodramus savannarum
Ammodramus humeralis
Ammodramus aurifrons
Rhynchospiza stolzmanni
Rhynchospiza strigiceps
Arremonops tocuyensis
Arremonops conirostris
Arremon taciturnus
Arremon semitorquatus
Arremon franciscanus
Arremon flavirostris
Arremon aurantiirostris
Arremon schlegeli
Arremon abeillei
Arremon brunneinucha
Arremon atricapillus
Arremon basilicus
Arremon perijanus
Arremon phaeopleurus
Arremon phygas
Arremon assimilis
Arremon torquatus
Arremon castaneiceps
Oreothraupis arremonops
Atlapetes albofrenatus
Atlapetes semirufus
Atlapetes personatus
Atlapetes albinucha
Atlapetes melanocephalus
Atlapetes pallidinucha
Atlapetes flaviceps
Atlapetes fuscoolivaceus
Atlapetes tricolor
Atlapetes leucopis
Atlapetes latinuchus
Atlapetes blancae
Atlapetes rufigenis
Atlapetes forbesi
Atlapetes melanopsis
Atlapetes schistaceus
Atlapetes leucopterus
Atlapetes albiceps
Atlapetes pallidiceps
Atlapetes seebohmi
Atlapetes nationi
Atlapetes canigenis
Atlapetes terborghi
Atlapetes melanolaemus
Atlapetes rufinucha
Atlapetes fulviceps
Atlapetes citrinellus
Chlorospingus flavopectus
Chlorospingus tacarcunae
Chlorospingus semifuscus
Chlorospingus parvirostris
Chlorospingus flavigularis
Chlorospingus flavovirens
Chlorospingus canigularis
New
information:
Klicka et al. (2014) produced the first comprehensive phylogeny for the family,
which they further restricted to New World genera under the name
Passerellidae. They found that the Emberiza buntings and relatives are not
the sister group to the New World sparrows (but that should be the topic of a
separate proposal, likely to go to NACC first).
The Klicka et al. phylogeny has exceptionally strong taxon sampling (at
least for mtDNA) and gene sampling (nuDNA sequenced for subset of critical
taxa), so I do not think we’ll be seeing anything more detailed any time
soon. The support values for most nodes
in the tree are strong:
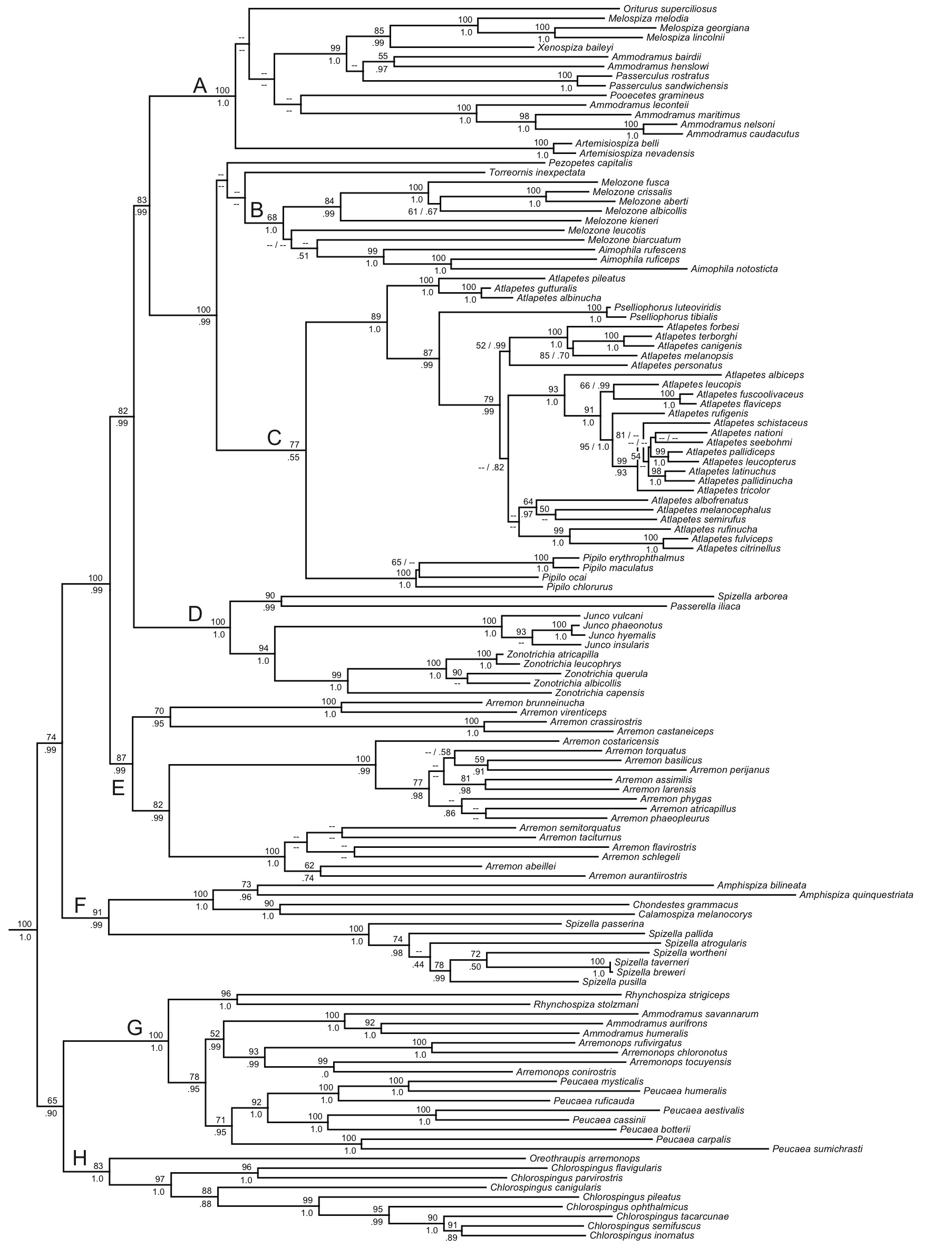
Using
our standard sequencing conventions, the following sequence reflects Klicka et
al.’s Fig. 1. I included all the taxa
restricted to North and Middle America (shaded gray) for completeness, and I
used indentation and skipped lines to help mimic the branching pattern. The one area of semi-exception to the
conventional rule is in Atlapetes. If you look at the tree, you can see that
there are many nodes with little or no support, and even so, Fig. 1 is based
entirely on mtDNA and thus may have gene tree/species tree problems. In fact, the *BEAST species tree in Fig. 2c
shows a large polytomy among the taxa sampled except for the northern group, of
which we only have A. albinucha. The
nuDNA-only tree (Fig. 2b) shows a similar pattern. Therefore, the proposed sequence, clearly
tentative, is a mix of incorporating the few solid nodes with geography, which
is clearly the best predictor of relationships in the genus, not color (as I
predicted in an Auk paper back in
1995 or so). The exception to geography as
a predictor of relationships would be if the Tepui species personatus is actually more closely related to a cluster of
narrowly distributed species in central/southern Peru, which would be very
interesting (and which has parallels, I think, in Myioborus and Myiothlypis).
Note
that Chlorospingus flavovirens is not in Fig. 1
- -this is because Klicka et al. (2014) found that not only is it not a Chlorospingus but it is a
member of the Thraupidae, not Emberizidae/Passerellidae. Until we have a genus for flavovirens,
and until we know where in the Thraupidae it goes, I suggest we leave it at the
end of Chlorospingus, with appropriate footnote.
One of the many
interesting points illuminated by the Klicka et al. (2014) that would be
reflected in the new sequence is that Chlorospingus is indeed embedded
in the family, but sister to everything except the enigmatic Oreothraupis. Another is that many of us might think of our
“northern” sparrows as a monophyletic group, but they are not. Also, the “brush-finches”, Atlapetes
and former Buarremon, are even more distantly related than was
recognized. (Extralimitally, note that
the Pselliophorus is really just an Atlapetes, and that Ammodramus
and Melozone are polyphyletic.)
Oreothraupis arremonops
Chlorospingus flavigularis
Chlorospingus parvirostris
Chlorospingus canigularis
Chlorospingus pileatus
Chlorospingus flavopectus
Chlorospingus tacarcunae
Chlorospingus
inornatus
Chlorospingus semifuscus
[Chlorospingus flavovirens – retained here temporarily pending additional
publication]
Rhynchospiza stolzmanni
Rhynchospiza strigiceps
Peucaea carpalis
Peucaea sumichrasti
Peucaea ruficauda
Peucaea humeralis
Peucaea mystacalis
Peucaea botterii
Peucaea cassinii
Peucaea aestivalis
Ammodramus savannarum
Ammodramus
humeralis
Ammodramus aurifrons
Arremonops
rufivirgatus
Arremonops
chloronotus
Arremonops conirostris
Arremonops
tocuyensis
“Spizella” arborea
Spizella passerina
Spizella pallida
Spizella atrogularis
Spizella pusilla
Spizella breweri
Spizella wortheni
Amphispiza
bilineata
Amphispiza
quinquestriata
Chondestes
grammacus
Calamospiza
melanocorys
Arremon costaricensis SS
Arremon basilicus SS
Arremon perijanus SS
Arremon atricapillus SS
Arremon phaeopleurus SS
Arremon phygas SS
Arremon assimilis SS
Arremon torquatus SS
Arremon aurantiirostris SS
Arremon abeillei SS
Arremon schlegeli SS
Arremon taciturnus SS
Arremon franciscanus * SS
Arremon semitorquatus SS
Arremon flavirostris SS
Arremon
virenticeps
Arremon brunneinucha
Arremon
crassirostris
Arremon castaneiceps
Passerella iliaca
Junco hyemalis SS
Junco phaeonotus SS
Junco vulcani SS
Zonotrichia capensis
Zonotrichia leucophrys
Zonotrichia atricapilla
Zonotrichia querula
Zonotrichia albicollis
Artemisiospiza
nevadensis
Artemisiospiza
belli
Oriturus superciliosus
Pooecetes gramineus
“Ammodramus”=Ammospiza
lecontei
“Ammodramus”=Ammospiza maritimus
“Ammodramus”=Ammospiza s nelsoni
“Ammodramus”=Ammospiza caudacutus
“Ammodramus”=Passerherbulus
bairdii
“Ammodramus”=Passerherbulus
henslowii
Passerculus
sandwichensis
Xenospiza baileyi
Melospiza melodia
Melospiza lincolnii
Melospiza georgiana
Pezopetes
capitalis
Torreornis inexpectata
Melozone kieneri
Melozone fusca
Melozone albicollis
Melozone aberti
Melozone crissalis
Melozone leucotis
Melozone biarcuata
Aimophila
rufescens
Aimophila ruficeps
Aimophila notosticta
Pipilo chlorurus
Pipilo maculatus SS
Pipilo
erythrophthalmus SS
Pipilo ocai SS
Atlapetes pileatus
Atlapetes albinucha
“Pselliophorus” tibialis
“Pselliophorus” luteoviridis
Atlapetes albofrenatus
Atlapetes personatus
Atlapetes melanocephalus
Atlapetes semirufus
Atlapetes flaviceps
Atlapetes fuscoolivaceus
Atlapetes leucopis
Atlapetes albiceps
Atlapetes rufigenis
Atlapetes tricolor
Atlapetes
schistaceus
Atlapetes
pallidinucha
Atlapetes
latinuchus
Atlapetes
leucopterus
Atlapetes
pallidiceps
Atlapetes
seebohmi
Atlapetes
nationi
Atlapetes forbesi
Atlapetes
melanopsis
Atlapetes
terborghi
Atlapetes
canigenis
Atlapetes melanolaemus*
Atlapetes rufinucha
Atlapetes
fulviceps
Atlapetes
citrinellus
Recommendation: The current sequence does not reflect the
most recent phylogenetic data and has to be changed. I encourage inspection of the tree and the
sequence to see whether tweaks are needed, especially in Atlapetes, which has severe problems with respect to placement of A. personatus and A. rufigenis if the sequence is to combine geography and phylogeny.
Van Remsen and John
Klicka, May 2014
________________________________________________________________________________________________________
Comments from Robbins: “YES, to the
sequence change based on the new molecular data.”
Comments from Stiles: “YES, to bring the sequence into line
with the phylogeny. Do we need a
separate proposal regarding splitting Passerellidae (New World) from the Old
World Emberizidae sensu stricto?
Comment from
Remsen in response to Stiles:
Yes we definitely need that proposal. I
was hoping that NACC would take the lead on this, but if anyone here wants to
go forward with one, do it.”
Comments from Nores: “? Although I agree that the current
sequence does not reflect the most recent phylogenetic data and has to be
changed, the new sequences (Arremon
and Atlapetes), in my opinion, need
tweaks.”
Comments
from Jaramillo: “YES
– This paper is a good one! Very well done. Apart from the various
considerations we are dealing with here, I was quite surprised to see were Torreornis fell out. Lots of unexpected
issues here. There may be some generic changes to come, I am not sure which
species carries the name Ammodramus
for example.”
Comments
from Pacheco:
“YES. Although the current sequence does
not reflect the most recent phylogenetic data, this suggestion to new linear
sequence is an improvement.”
Comments
from Zimmer: “YES. Not perfect, but the new linear sequence does
reflect the most recent molecular data, and represents a step forward.”
Comments
from Pérez-Emán: “YES
as this new linear sequence incorporates the most recent information
(molecular) on the group. About A.
personatus, it should be grouped together with central/southern Peruvian
species. A study on which John, Daniel and I has been working for a while,
including a more complete sampling for the genus, supports the relationship of
this species with Peruvian species.”