Proposal (1008) to South
American Classification Committee
Treat Tunchiornis ochraceiceps
(Tawny-crowned Greenlet) as consisting of four species
Effect
on SACC list: This proposal would treat our current Tunchiornis
ochraceiceps as consisting of four species.
Background: Our current Note reads as follows:
14a. See Ridgely & Tudor (1989) for potential reasons for ranking of the southern rubrifrons subspecies group as a separate species from Hylophilus ochraceiceps. Slager et al. (2014) found deep divergences within among lineages included in H. ochraceiceps. Del Hoyo & Collar (2016) treated luteifrons of the Guianan Shield as a separate species (“Olive-crowned Greenlet”) based in part on Slager et al. (2014) and also on vocal differences pointed out by Boesman (2016h). Buainain et al. (2021) found evidence for treating it as consisting at four species; they found that luteifrons is sister to the rubrifrons group of southeastern Amazonia, treated as conspecific with ochraceiceps by del Hoyo & Collar (2016). SACC proposal badly needed.
Tunchiornis
ochraceiceps
(Tawny-crowned Greenlet) is a polytypic species with a large distribution in
the Neotropics; 10 subspecies occur from s. Mexico to southern Amazonia. They have always been treated as part of the
same species (as far as I know), e.g., Hellmayr 1936, Blake 1968 “Peters”,
Meyer de Schauensee 1970). The only
major break in the distribution is across the Andes, with the number of
subspecies equally divided between trans-Andean and cis-Andean populations. It is treated as a single species by
Dickinson & Christidis (2014) and IOC (v. 13.2; 2023).
Boesman
(2016) sampled 6 recordings of trans-Andean ochraceiceps, 9 of the
Amazonian ferrugineifrons-rubrifrons group, and 8 of Guianan
Shield luteifrons. Unfortunately,
the locations and subspecies allocations were not reported, so for example
there is no way to know which Amazonian taxa were sampled, which is critical
given that Buainain et al. considered ferrugineifrons and rubrifrons
to be separate species. Boesman found
that luteifrons differed strongly from his other two groups: “Surprisingly, the different song of luteifrons
has seemingly nowhere been picked up in literature: its song consists of two
notes with decreasing pitch (score 4), resulting in an overall longer song
phrase (score 2-3) and larger frequency range (score 1-2). When applying Tobias
criteria, this would lead to a total vocal score of about 5 vs. all other races.”
And “All in all, we can conclude that the Guianan group
clearly stands apart vocally.”
Here's
the map (from Buainain et al. 2021), which will be useful in evaluating the
proposal:
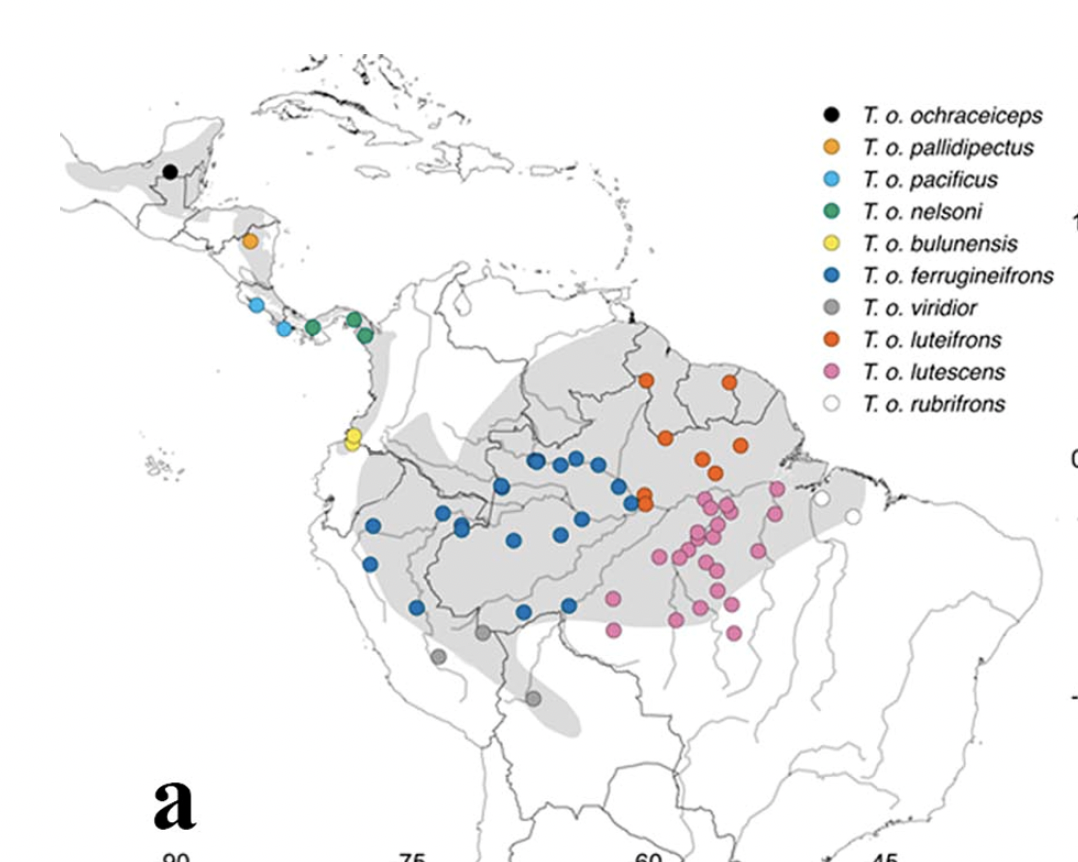
New
information:
Nelson Buainain and colleagues used 625 specimens, 152 vocal recordings, and 69
tissue samples, with complete taxonomic and broad geographic sampling. The genetic results were based on UCE data
from 2267 usable loci. The vocal results
were based on analysis of a typical range of characters quantified from
sonograms. Standard morphometric
measurements were used to evaluate morphological differences. Plumage variation was analyzed by applying
Smithe color names and also by simultaneous comparisons of a large series of
specimens assembled at the AMNH; photographs of all type specimens were
examined as well as all original descriptions.
This is another outstanding empirical study coming out of Brazil.
A
detailed synopsis of all these data would take too much space here. The analyses are detailed and objective. My favorite is the Estimated Effective
Migration Surface analysis in Fig. 3 – cool stuff. The basic point that
this bird consists of a bunch of old lineages with minimal phenotypic
differentiation is biologically important, and as analyses of Neotropical
lineages in this detail increase, through a comparative framework perhaps we
will gain a better idea for why some change quickly whereas others do not. If I’d reviewed the paper, I would have
pointed out that one of Joel’s own papers could have been used to illustrate
conservative phenotypic evolution in the group: Reddy & Cracraft (2007; MPE
44: 1352-1357) showed that two Indomalayan genera are actually sister New World
vireos, and in fact Indomalayan Erpornis looks remarkably like a Hylophilus
sensu lato despite ca. 30 MY of separation, and some Indomalayan Pteruthius
resemble Vireolanius. Anyway, congrats
to the authors on a great paper. Check
out the details for yourselves. My only
criticism is that when they listed their four species, a Diagnosis for each,
summarizing their data, would have been very useful. As is, one has to backtrack through the paper
to piece together the characters used to delimit the four species-level taxa
(and two additional subspecies-level taxa).
From
the standpoint of taxonomy, here’s what stands out to me:
Plumage: Of consequence to
eventually incorporating subspecies taxonomy into SACC, only two of the five trans-Andean
subspecies is 100% diagnosable. The plumage
variation is almost all clinal, from s. Mexico to nw. Ecuador, with a blip due
to Gloger’s Rule, according to their analyses.
However, no official tests of diagnosability were conducted on the
plumage data, which constitutes the basis of the subspecies original
descriptions; rather than simply point out that they were not diagnosable, perhaps
a better approach would have been to show that quantitatively to see what the
level of diagnosability is because even the most rigid application of PSC-like
thinking allows for something below 100% diagnosability. Before synonymizing all 5 subspecies, the
data should be re-evaluated from this standpoint; the pattern is clearly
clinal, but is it a step cline, with each of the 5 subspecies representing
plateaus? The subspecies nelsoni is
stated as intermediate between northern ochraceiceps and bulunensis of the Chocó, and is also clearly a
genetically admixed population of the two in terms of mtDNA, but is it a
phenotypically diagnosable unit? The
text mentions differences in iris color between some groups, but apparently
these are not fixed.
Three subspecies groups are, however,
diagnosable by head coloration, and these are two of the taxa that they
proposed to be elevated to species rank (see below): T. luteifrons of
the Guianan Shield and T. rubrifrons of SE Amazonia. However, they evidently could not diagnose
the Middle American ochraceiceps group from ferrugineifrons from
W Amazonia, although they stated that there are trends in coloration that
correspond to these groupings. Thus, it
is also not clear to me now whether the two subspecies taxa retained in their
classification are diagnosable only as mtDNA lineages, in which case there is
no basis for recognition as subspecies, in my opinion. I’m confused.
Perhaps all of this explained and detailed in
Supplementary Information, although there is no text reference to such files on
morphology. For some reason, I cannot
access the SI.
Morphometrics: None of the taxa,
even those ranked as species by the authors, is diagnosable by
morphometrics. No surprise there.
Vocalizations: Vocalizations of
course are critical to species limits in the Vireonidae. As far as I know, all taxa recognized as
species in the family differ in vocalizations.
For several decades, I’ve heard through others that vocal differences were
notable among some of the populations of T. ochraceiceps. Therefore, I was expecting their analysis of
150+ recordings to confirm the discrete differences I had heard discussed. Here is their lead sentence:
“The
qualitative analysis shows no diagnostic vocal characters for any of the
populations of T. ochraceiceps (Fig. 8a–d). Some variation, however, is
noteworthy and a relatively reliable indicator of population assignment.”
They
provided the following comments on the vocal differences among their geographic
groups, for which I am adding their proposed species name rather than the
geographic clusters in the paper:
•
ochraceiceps: “The trans-Andean
forms tend to have higher frequency (higher pitch) songs (Fig. 9c-g), although
there is much overlap with other populations.”
•
luteifrons: “The NEA populations
east of the Branco River, predominantly produced a distinct song with two
whistled syllables, between the frequencies of ~ 2.7–3.8 KHz,
being the last syllable descending in modulation. However, it is clear that
these birds can eventually produce one syllable vocalizations (ML 80429, NB
observation during field work around Manaus, AM, Brazil) that are apparently
indistinguishable from songs with one syllable from the other populations.”
•
rubrifrons: “All recordings from
the Madeira-Tapajos ´ interfluve, in SEA, contained a distinct song with two to
three syllables, between the frequencies 2.8–3.5 KHz,
but with ascending second and third syllables, thus different from the NEA
population in modulation. However, songs with one and two syllables were
recorded in the Juruena-Teles Pires (whose confluence form the Tapajos River)
interfluve. This area lies in the limit between the single and multiple
syllable song populations in SEA. Thus, although some interesting difference in
frequency of vocal pattern exist, it is not possible to unmistakably
distinguish populations by their songs.”
•
ferrugineifrons: [not really discussed per se, as far as I can tell, but
see graph below]
Here
is their Fig. 9h, which is a PCA classification of quantitative vocal
characters:
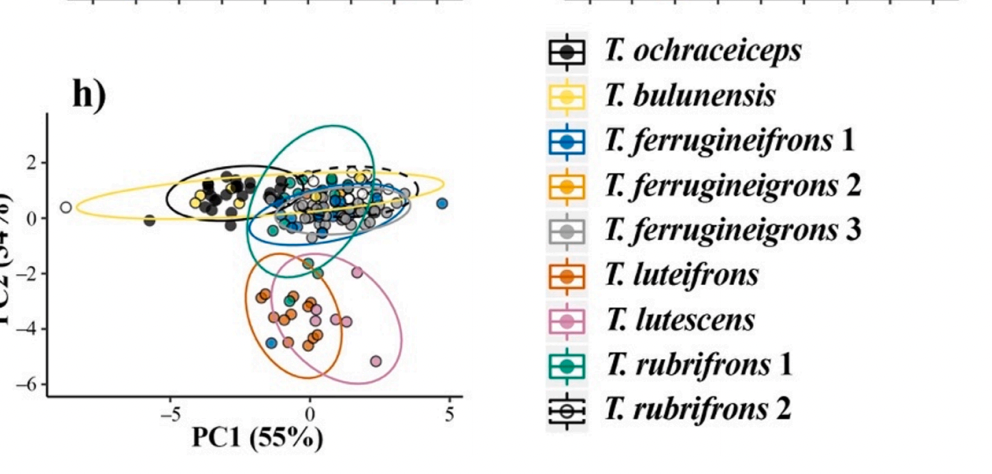
I
find this figure difficult to decipher, but what stands out to mw is that there
are 3 clusters: (1) upper left, which corresponds mainly to the nominate
ochraceiceps group, but also some portion of the Chocó taxon bulunensis (accidentally treated as a species
in the legend); upper right, which is a supo of rubrifrons and ferrugineifrons
1, “ferrugineigrons” 3 [unfortunately conspicuous
typo missed by 8 authors, and an unknown number of editors and reviewers], and
some bulunensis; perhaps due to poor eyesight,
I cannot spot where T. “ferrugineigrons” 2 is;
and (3) T. luteifrons and what is marked as T. lutescens, which
they consider a subspecies of T. rubrifrons in their
classification. Thus, I really don’t
know what to make of all this. As noted
in their lead sentence above, the authors stated upfront that none of the
populations have diagnostic vocal characters.
Certainly, as far as I can tell, vocal differences seem somewhat chaotic
and don’t map well on to their genetic groups.
I hope someone else can dig further into this in case I am
misinterpreting something.
With
all appropriate caveats from concluding anything from single recordings, here
are a few I picked out from xeno-canto.
This is tedious because the number of recordists who rate their
recordings as “A” quality have obviously never listened to, for example, an
Andrew Spencer recording among others, so you have to wade through lot of
mediocre recordings to get to a good one.
I think xeno-canto is one of the world’s great bird resources; I just
wish there was a more objective way to rate recordings other than self-rating,
so that the truly best would come nearest the top.
• ochraceiceps (from Puntarenas, Costa
Rica, by Peter Boesman): https://xeno-canto.org/274157
• luteifrons (from Brownsberg,
Suriname, by Peter Boesman): https://xeno-canto.org/271834
• ferrugineifrons (Rîo
Javarí, Peru, by Peter Boesman) https://xeno-canto.org/270735
• rubrifrons (Cristalino
Lodge, n. Mato Grosso, Brazil, by Frank Lambert): https://xeno-canto.org/68496
From
the standpoint of someone who was familiar (once upon a time) only with ferrugineifrons
from Bolivia, these all basically sound similar to someone not embedded in the
greenlet voice universe; even the two-noted luteifrons doesn’t sound
radically different from the others.
Genetic
divergence:
Here is their main figure. I have superimposed the four proposed species
name in blue on each cluster. Obviously T.
ochraceifrons consist of four fairly old lineages in terms of
mtDNA. The estimated divergence time of
the ochraceiceps cluster from the ferrugineifrons cluster is
almost 5 MYA (Pliocene), which is older than many crown lineages we rank as
species. The separation of the crown
lineages luteifrons and rubrifrons is much older, ca. 7.5. MYA
(Miocene), i.e. older than many crown lineages we rank at the genus level. I do not have the qualifications to evaluate
their time calibrations, but taken at face value, they certainly are consistent
with species rank.
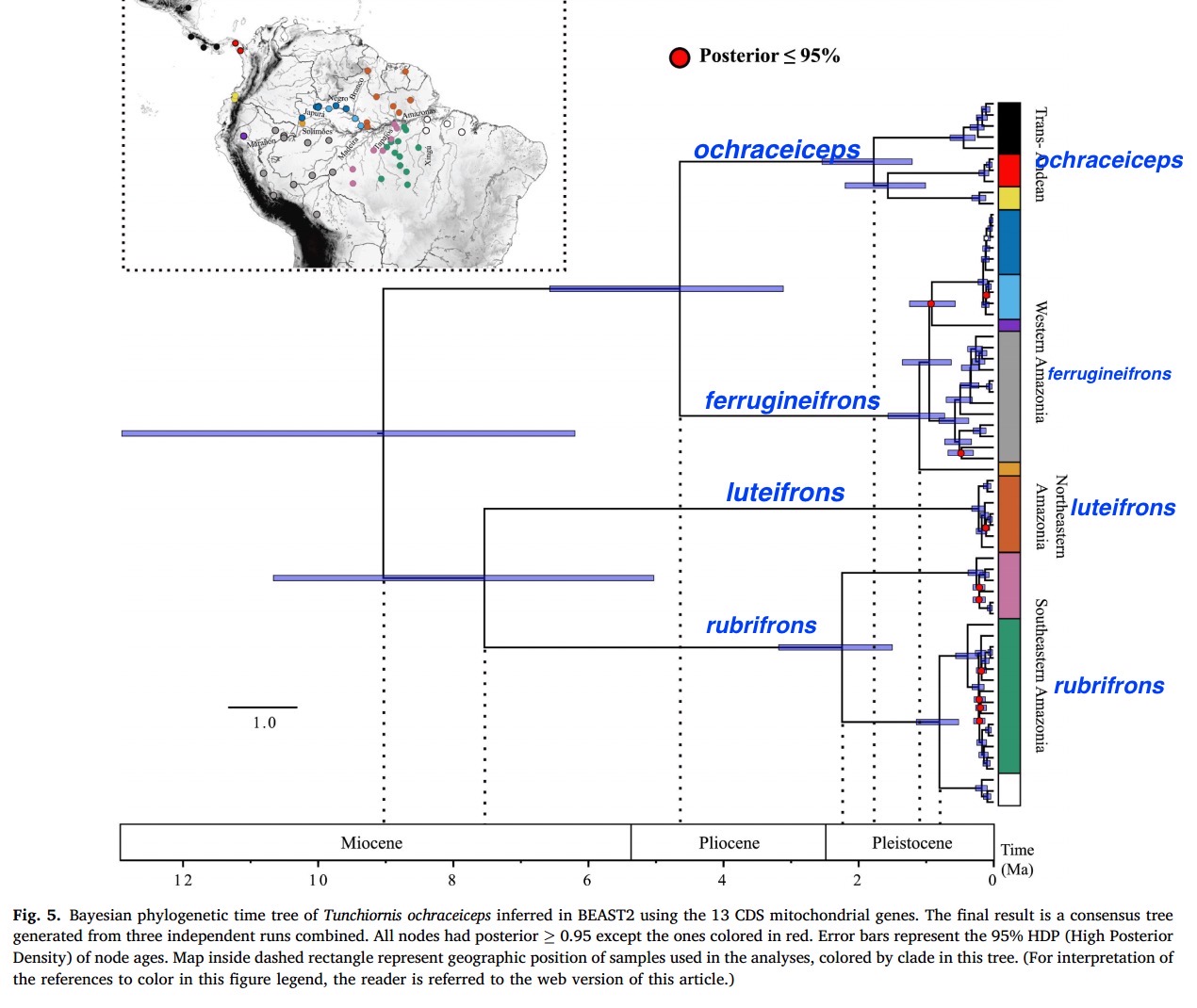
A
summary of their recommended species classification is as follows:
• Tunchiornis
ochraceiceps:
s. Mexico through Central America to nw. Colombia and south in Chocó region to
nw. Ecuador; includes T. o. bulunensis.
• Tunchiornis
ferrugineifrons
(Sclater, 1862): w. Amazonia, from W of Rio Branco through s. Venezuela,
se. Colombia, e. Ecuador, e. Peru, and ne. Bolivia to w. bank of Rio Madeira in
sw. Amazonian Brazil.
• Tunchiornis
luteifrons
(Sclater, 1891): Guianan Shield east of Rio Branco through the Guianas to n. Pará and Amapá to n. bank
of Amazon.
•
Tunchiornis rubrifrons (Sclater & Salvin, 1867): s. Amazonia S of the Amazon from
Rio Madeira east to Rio Tocantins and se. Pará (?and w. Maranhão), including T.
r. lutescens)
Here
is their figure with geographic distributions and subspecies/species
classification
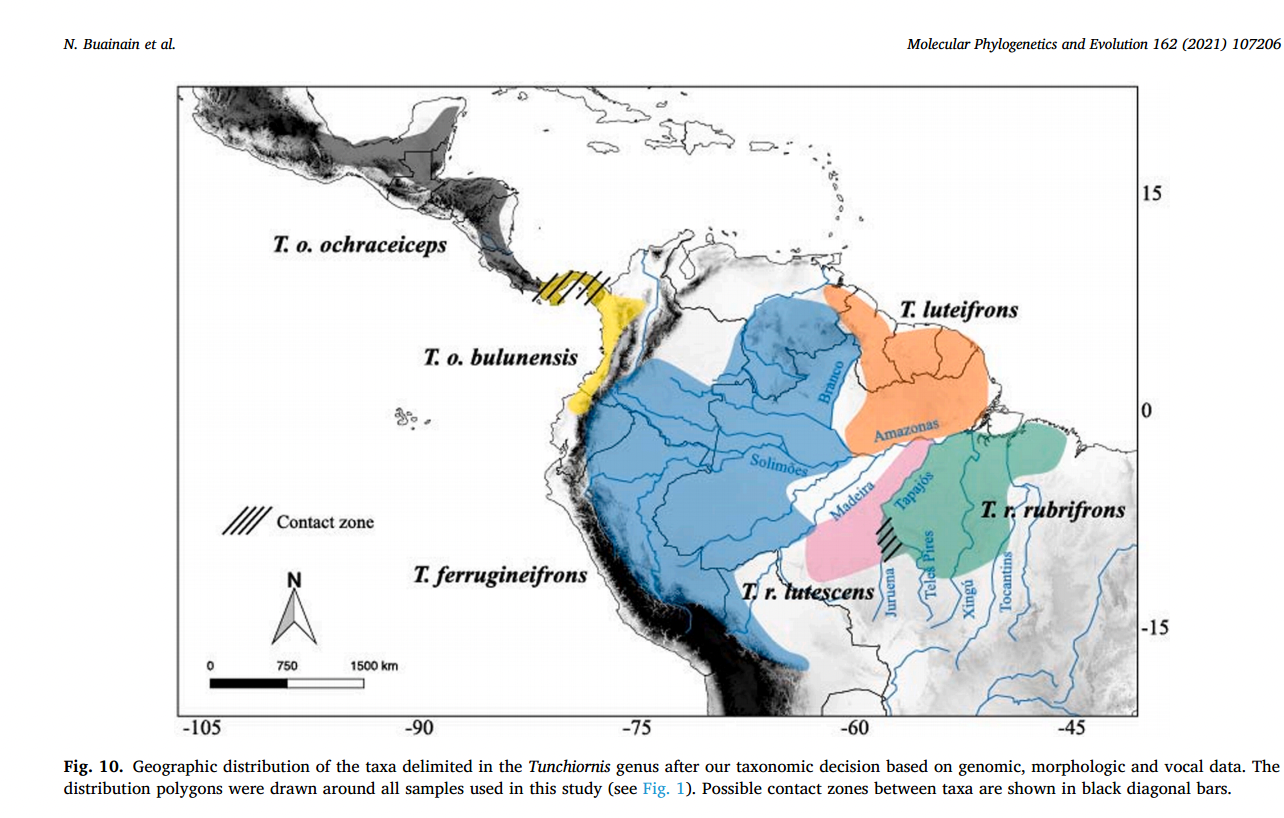
Discussion: Basically, my impression is that the data are
terrific, but the interpretations of the analyses are insufficiently clear, at
least to me, for sorting it all out in terms of taxonomy. The genetic results scream out for
recognition of at least four species, but when one tries to diagnose these
vocally, it falls apart, with a few instances of songs mismatched to genetic
group and no crystal clear vocal groups.
I suspect a reanalysis of plumage characters would reveal not just three
but at least four and probably many more phenotypically diagnosable units. Central to the problem is that luteifrons
appears to be the most distinctive taxon in terms of both voice and plumage (no
rufous color on crown or face), yet it is sister to rubrifrons, which
cannot be separated, evidently, from ferrugineifrons vocally. So, what we have here is a case of unequal
rates of character evolution, which poses a dilemma for species concepts.
Biologically, this is a fascinating
situation. Genetically, the results seem
crystal clear and tidy, but once one tries to integrate this with plumage and
voice, it’s a mess. The authors say as
much in the paper. Do we go with the
deep genetic divergence and ignore apparent lack of phenotypic
distinctiveness? Vireonidae has may
cases of barely differentiated taxa treated as species, e.g. most recently Vireo
chivi and V. olivaceus.
I have no recommendation either way –
I’m going to see what others say, especially with regard to voice.
For
voting, let’s do it this way:
YES
= Recognize
4 species, as per recommendations in the paper.
NO
= Do
not recognize 4 species at this point but maintain status quo, at least
temporarily, with acknowledgement that the complex may contain 1, 2, 3, or 4
species pending further evaluation.
English
names: If the proposal passes, then we will need a
separate proposal on English names,
References:
Boesman, P.F.D. 2016.
Notes on the vocalizations of Tawny-crowned Greenlet (Hylophilus ochraceiceps). Ornithological Note 168. Birds of the World, Cornell Lab of
Ornithology.
BUAINAIN, N., M. F.A.
MAXIMIANO, M. FERREIRA, A. ALEIXO, B. C. FAIRCLOTH, R. T. BRUMFIELD, J.
CRACRAFT, AND C. C. RIBAS. 2021. Multiple species and deep genomic divergences
despite little phenotypic differentiation in an ancient Neotropical songbird, Tunchiornis
ochraceiceps (Sclater, 1860) (Aves: Vireonidae). Molecular Phylogenetics Evolution 162:
107206.
Van Remsen, September
2021 rev. June 2024
Vote tracking chart:
https://www.museum.lsu.edu/~Remsen/SACCPropChart968-1043.htm
Comments
from Areta:
“YES. I vote for the 4-way split. The
deep genetic differences coupled with somewhat different vocalizations,
plumages and eye-color, indicate that keeping all these taxa as a single
species is incorrect. I share Van´s misgivings on how the information is
organized and summarized, and I think that more detailed vocal analyses will
uncover more clear vocal distinctions among the four main species-level taxa.”
Comments
from Claramunt:
“YES. I think the proposed taxonomy is a step
forward. The data clearly show that there are multiple species-level lineages
in this complex and the proposed splits are supported by evidence.”
Comments
from Robbins:
“Unless one strictly relies on the mtDNA for making species allocations, this
is messy from a vocal and plumage morphology standpoint. An example of vocal
issues, I have recorded birds in Nicaragua and Guyana that both give single and
two-noted calls that are very similar. Listening to recordings from elsewhere I
get the same impression as what is stated in the paper, seems to be no
diagnosable song for each of the four clades.
As Van pointed out, conservative plumage morphology may be clinal and it
is unclear whether there are diagnosable characters for each clade. Unless I
missed something, it is unclear if there are consistent differences in eye
color. So, I’m on the fence on this one and look forward to seeing comments by
others.”
Comments
from Jaramillo:
“YES – This is a complicated group with lack of outstanding plumage features to
work with. I guess this is why they call them greenlets. In any case, I am
persuaded by the data here and the summary that Van provides. All Vireonidae
differ in vocalizations – Van have you heard singing Philadelphia Vireos? Even
the Red-eyes can’t tell them apart!! But yes, their calls are different.”
Comments
from Stiles:
“YES. The 4-way split of Tunchiornis seems well justified – it certainly
is a long step forward in understanding Tunchiornis and serves as a
solid base should additional data indicate further splitting in the future.”
Comments
from Lane:
“NO. Ugh. This is one of those cases that just doesn’t really feel like it is
cooked enough to take out of the oven. I guess we can just take the molecular
results and shrug at the fact that the plumage and voice data don’t map well
onto them. I just did a quick search as per Alvaro’s comments to see if there
are enough recordings of calls to provide support in that group of
vocalizations being stronger evidence for differences, but there are very few
call recordings (at least in X-c) for several of the
taxonomic groups so it is hard to say. I really would rather wait for more
evidence to elucidate the situation before accepting splits here. NO.”
Comments
from Andrew Spencer (who has Remsen vote): “YES - this was a really tough vote
for me. No one factor convinces me that these should be split, but when taken
together they tip me into the yes camp. Regarding vocalizations, as others
have already stated here, the situation seems really murky. Listening
to recordings, I do get the general patterns laid out by Boesman and expanded
on by Buainain, especially regarding luteifrons and also trans-Andean
birds vs. the rest. But the variation in any one group does muddy the picture
significantly. That said, I don't particularly see that as a mark against a
split. Philadelphia and Red-eyed vireos songs are often completely
indistinguishable by anyone (including themselves), and others like some
members of the former Solitary Vireo differ mostly in average characters
somewhat like these greenlets. Conversely, vocal differences by themselves
aren't going to persuade me to split these or other Vireonidae, given
dialectical differences in other members of the family that (in my
opinion) aren't indicative of separate species status. Calls may well be a
better avenue to show speciation here, but even if they eventually prove to not
be that different I still think my points above stand.
“It's
those average differences combined with the other evidence presented persuades
me to still vote for the split. I understand Dan's point that this has some
aspects of "doesn’t really feel like it is cooked enough", to use his
terminology. But I do think that the four species interpretation is a step
forward in our understanding of the complex, and in my opinion at least, a
significant step forward.”
Comments
from Mario Cohn-Haft (who has Del-Rio vote): “YES. I basically agree with everybody
else (the nays and yays). Here in the central Amazon,
it's really obvious that there are consistently distinct voices and plumages
across the rios Negro and Madeira and lower Amazon,
and seemingly across the Tapajos as well. So, the 4-way split proposed is, I would say,
a conservative description of reality (there are probably more than 4 spp.
involved) and a perfectly reasonable conclusion from the point of view of
nomenclature and distributions.
“On
the other hand, the story as published leaves a lot to be desired in the way of
convincing evidence. That is, I'm more convinced by my own experience
than by what's available in the literature. And I think the reason for that is that a
proper vocal analysis requires a lot more work and thought than anybody has
been willing to give it so far. I'm the first to admit that I didn't tackle the
problem myself out of sheer laziness. Way back in the late 80s when Ridgely's
frustratingly erroneous description of the situation irked me into actually
thinking about doing something, I realized what a big job it was going to be,
and I've sat around hoping someone would do it up right ever since. Yes, some populations can produce
vocalizations just like others, making 100% diagnosis on one character alone
difficult. So what? (Ah, and guess what, that happens with
suboscines too.) A good vocal analysis needs to take into account repertoire,
context of vocalizations, frequency (not kHz, but opposite of rarity--the number
of times that a particular vocalization is given); carefully chosen vocal
variables (not a universal recipe of maxs, mins,
etc.-- I could go on and on about what good vocal variables are, but this isn't
the place) and comparison of analogous vocalizations, not just everything at
random; means and variances, for example, only make sense in these specific
contexts. What we're seeing consistently nowadays are molecular studies to
which vocal or morphological components are added almost as an afterthought to
"strengthen" (or not) the molecular conclusion, but that do not stand
on their own. In that sense I totally agree with Dan and others who say this
cake is only half-baked at best. (By the way, I can't remember if iris color
entered into any of this, but it's also a relevant diagnostic character in this
complex.)
“The
hard question for me is: How purist (perfectionist?) are we going to be? I
think the 4-spp answer is right (and an underestimate), so how much longer will
we wait before it makes it into the checklist? If we were Peters, or better yet
my hero John Zimmer, we'd just declare it and be done with it and let
nay-sayers or future generations of topic-challenged grad students worry about
the details. And that's what I'm inclined to do. I vote YES, believing it's the correct answer,
while recognizing that the published and available evidence don't tell a
thoroughly convincing story. Messy? A bit authoritative or arbitrary? Sorry.
Watching the planet burn, flood, become bare of vegetation at the rate it is
now, I find it hard to justify waiting to do up properly the taxonomy of what I
used to hold aside as "my pristine Amazon". We are literally on the
brink of cataloging extinct species.”
Comments
from Zimmer:
“YES. A complicated one
indeed. The depth of the genetic splits
here are impressive, and combined with some average differences in plumage,
iris color, and vocalizations, even in the apparent absence of diagnosability,
are enough to sway me. As Andrew and
Alvaro both note, songs of some accepted species pairs of vireos (Red-eyed vs.
Philadelphia; all 3 of the splits from former “Solitary” Vireo) are doubtfully
diagnostic. I will say, that without
having ever conducted anything approaching a quantitative analysis, I’ve long
been struck by apparent vocal differences corresponding to each of these
suggested four species. I agree with
Mark that some populations can give either 1 or 2-note songs, and that they are
all broadly similar in quality, but nonetheless, I can still hear qualitative
differences in inflection and pitch.
Granted, these may not hold up to broad geographic sampling of each
population, but, at least in my coarse-grained field experience with this
group, it has always been my impression that there were some pretty consistent
geographic differences in songs that correspond pretty well to the partitioning
suggested by the genetic data. The case
is a long way from perfect, but I would agree with Andrew that this represents
a step forward in our understanding, and I think the genetic data combined with
the sum of mean differences in various character states is enough to hang our
hats on.”
Comments
from Bonaccorso:
“YES. Although the plumage differences are not entirely diagnosable, and the
song data are limited and substantially overlapping, neither type of character
alone is expected to be diagnostic for this group. However, the molecular
divergences, even if based only on mitochondrial DNA, are impressive.”
Additional
comments from Robbins:
“Given that the vast majority supported recognition of the 4 way split, I will
shift my on the fence vote to a YES.”
Comments
from Naka:
“YES. As far as I understand it, the 4-spp split fixes most problems from the
Brazilian Amazon, particularly within the Guiana Shield, where luteifrons
and ferrugineifrons replace one another. Along the Rio Branco suture
zone, birds are genetically, vocally, and morphologically distinct, which
includes a pale iris in ferrugineifrons, as Mario mentioned on his vote.
So, despite some concerns, I agree with Kevin in that those split populations
have always looked and sounded different to me.”