Proposal (#382) to South American Classification Committee
Change linear sequence of genera in the
Cotingidae
This proposal is to
change the current linear sequence of the currently recognized genera in the
Cotingidae to reflect recent phylogenetic data (Ohlson et al. 2007).
Background: Currently, the SACC sequence is:
Ampelion
Zaratornis
Doliornis
Phytotoma
Carpornis
Pipreola
Ampelioides
Rupicola
Phoenicircus
Cotinga
Procnias
Tijuca
Lipaugus
Conioptilon
Snowornis
Porphyrolaema
Xipholena
Carpodectes
Gymnoderus
Querula
Haematoderus
Pyroderus
Perissocephalus
Cephalopterus
This sequence
attempted to integrate the phylogenetic data in Prum & Lanyon (1989), Prum
(1990a, 2001), and Prum et al. (2000).
New data:
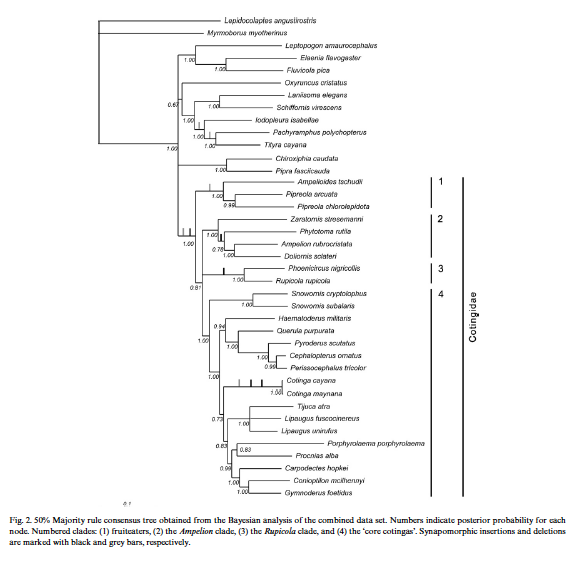
Ohlson et al. (2007) sequenced DNA from members of all genera except Carpornis; gene regions sampled were
included nuclear (myoglobin intron-2 and G3PDH intron-11) and mitochondrial DNA
(Cyt-b). The aligned sequences totaled
2118 bp. Their tree had good Bayesian
support at almost all nodes for terminal branches as well as at many interior
nodes. Among the latter, with 1.00 to
0.99 Bayesian support, were (1) Cotingidae as we currently define it as
monophyletic; (2) a group that includes Zaratornis,
Phytotoma, Doliornis, and Ampelion;
(3) a group that includes Pipreola +Ampelioides; a group that includes Snowornis, Haematoderus, Querula, Pyroderus, Cephalopterus, Perissocephalus,
Cotinga, Tijuca, Lipaugus, Porphyrolaema, Procnias, Carpodectes, Conioptilon, and Gymnoderus; (4) everything in 3 except Snowornis; (5) and Querula,
Pyroderus, Cephalopterus, and Perissocephalus.
Translating that to
a linear sequence, using the usual conventions (“basal” taxa first; for sister
taxa, NW-most taxon listed first; for polytomies, stay as close to traditional
sequence as possible), the result is shown below, with well-supported groups
color-coded, and indentations attempting to mark branching patterns within each
group. Within each grouping of more than
two taxa, some rotation is possible, and I have tried to stick to conventions
above for determining which comes first:
Pipreola
Ampelioides
Zaratornis
Phytotoma
Doliornis
Ampelion
Phoenicircus
Rupicola
Snowornis
Haematoderus
Querula
Pyroderus
Cephalopterus
Perissocephalus
Cotinga
Lipaugus
Tijuca
Procnias
Porphyrolaema
Carpodectes
Xipholena
Gymnoderus
Conioptilon
This omits Carpornis, for which there are no
data. However, Carpornis shows sufficient superficial similarities to Pipreola that it has been traditionally
placed adjacent to it. If we place it first
in the sequence, thus next to Pipreola,
we can maintain that historical tradition and add a footnote on the absence of
data. Placing it last to indicate
Incertae Sedis position is another option, but placing it first is another to
indicate I.S.
Analysis
and Recommendation: Linear sequences intuitively frame the way
researchers think about species and should reflect our best data on
relationships. A number of slight
permutations on this one are possible – if anyone sees improvements, these can
be discussed and worked in. Regardless,
the basic sequence above reflects our understanding of relationships within the
family much better than the previous one, and so I recommend a YES on this.
Lit Cit
OHLSON, J. I.,
R. O. PRUM, AND P. G. P. ERICSON. 2007. A molecular phylogeny of the cotingas
(Aves: Cotingidae). Molecular Phylogenetics and Evolution 42: 25-37.
Van Remsen (in consultation with Jan
Ohlson and Rick Prum, and input from Manuel Nores)
Comments
from Stiles:
“YES. Again, this study, involving both mitochondrial and nuclear genes and
with morphological input, clearly presents the best available information on
relationships to date. Classifying this family by plumage data or displays is
especially problematic because so many species form leks and thus are likely
under strong sexual selection, which often produces striking and unpredictable
divergences in such characters between related species.”
Comments
from Nores:
“YES. El nuevo ordenamiento
luce mejor, ya que está de acuerdo con la secuencia molecular de Ohlson y Prum.”
Comments
from Jaramillo:
“YES – Another straightforward and well-grounded proposal. I agree with the
suggestion to put Carpornis next to Pipreola and at the start of the
sequence.”
Comments from Zimmer:
“YES. This arrangement is in keeping with the best
and most recent available evidence. I
would favor placing Carpornis at the
beginning of the sequence to indicate incertae
sedis, not to indicate any particular relationship with Pipreola. There are some obvious plumage similarities
between Pipreola and Carpornis, but vocalizations couldn’t be
more different, with the two species of Carpornis
having loud, arresting vocalizations that cover great distances (when cucullatus is breeding, its song is
arguably the dominant voice of the forest, unless of course, Procnias is drowning it out.), whereas
every Pipreola I’ve ever heard has an
easily overlooked very thin, high-frequency, long whistle or trill and some
equally thin staccato chatter.
Morphologically, most Pipreola
exhibit a greater degree of sexual dimorphism, and all species in the genus
have a brightly colored orange/red bill.
Carpornis shows less sexual
dimorphism, and both sexes of both species have black bills. Pipreola
is, I think without exception, an exclusively montane genus (with a few species
found in the tropical zone of the foothills), and almost entirely Andean in
distribution (except for whitelyi of
the Guianan highlands). I tend to see
them mostly in the company of mixed-species flocks of frugivores, and often in
small groups consisting of several individuals of their own species. The two Carpornis
on the other hand, are endemics of Brazil’s Atlantic Forest, and although cucullatus is a foothill bird, melanocephalus ranges down to sea
level. They are generally solitary birds
(encountered as pairs or individuals, but not regularly in small groups), and
are not generally found with mixed-species flocks.”
Comments from Pacheco:
"YES. A sequência
sugerida é consistente com a mais recentes evidências. Concordo com Kevin que
um alegado parentesco de Carpornis com Pipreola não encontra um
reforço apropriado a partir das informações biológicas disponíveis.”