Proposal (591) to South American Classification Committee
Revise
the classification of the Pipridae
Effect
on SACC:
If adopted, this would (A) recognize the newly named genus Cryptopipo for Xenopipo holochlora,
(B) revise the linear sequence of genera in the family, and (C) recognize two
subfamilies within the family.
Background: Our current
classification of the Pipridae reflects substantial new increments of data from
recent research. Our note summarizes the
situation as follows:
1.
Sequence of genera and composition of the family follow Prum (1990a, 1992).
Genetic data (Tello et al. 2009, McKay et al. 2010) confirm the monophyly of
the Pipridae as constituted above, but also provide evidence that the existing
linear sequence of genera conflicts in several ways with phylogenetic
data. SACC proposal passed to change linear sequence of genera. Ohlson et al. (2013) proposed recognizing two
subfamilies, Neopelminae and Piprinae, and additional tribes within
Piprinae. SACC
proposal needed; their results also indicate that modifications to the
linear sequence of genera are also needed.
SACC proposal needed.
New
information:
Ohlson et al. (2013) investigated relationships
within the family using DNA sequence data from three nuclear introns and one
mitochondrial gene (ND2). They sampled
all genera and most species. I have
pasted in a screen grab of their tree below:
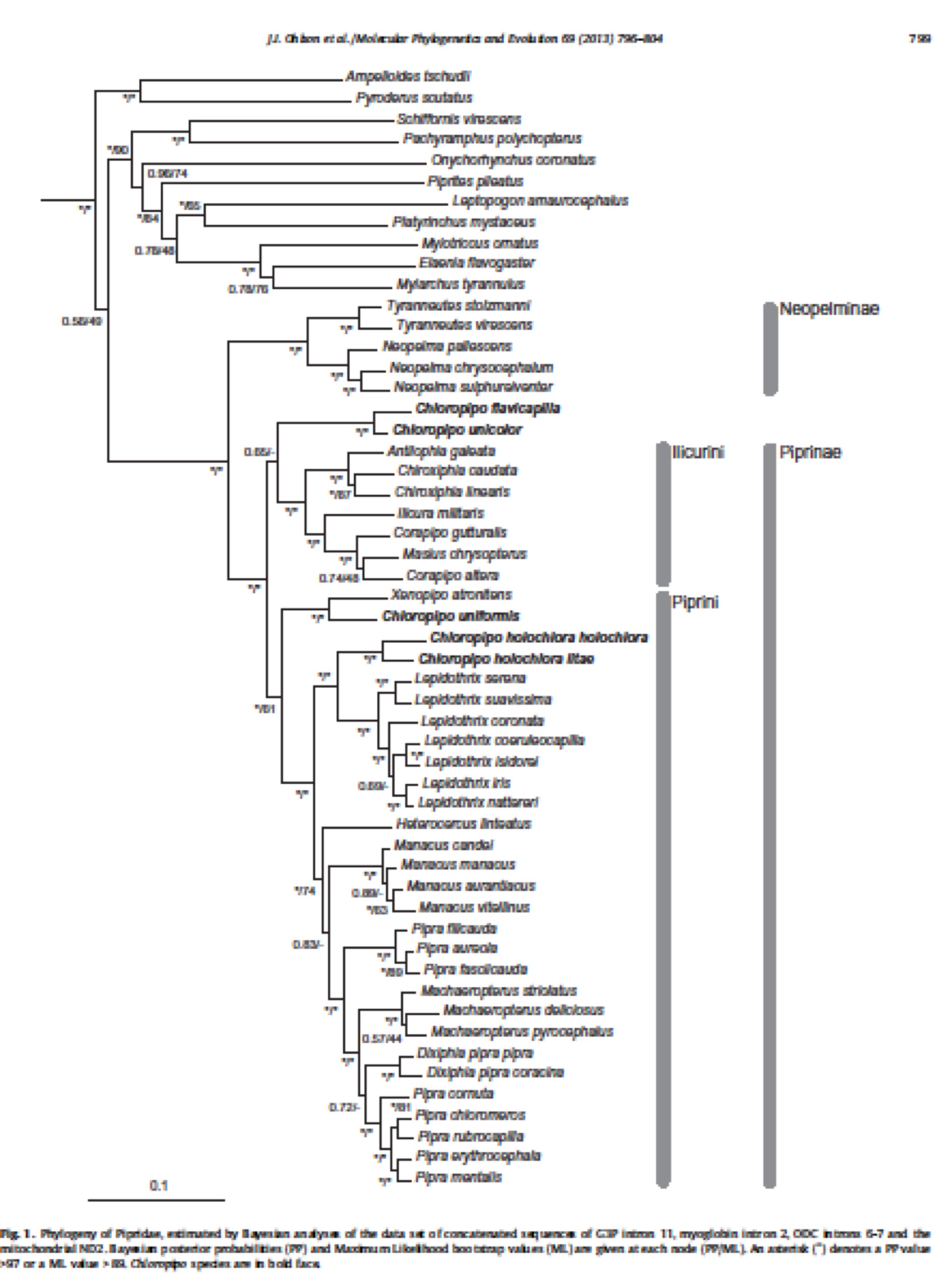
Their results are largely consistent
with those of previous studies except for the polyphyly of Chloropipo, members of which are in three parts of the tree. Because Ohlson et al. was the first study to
include all five species in broadly defined Xenopipo,
these results do not conflict with previous studies. McKay et al. (2010) included only unicolor, and Tello et al. (2009)
included only atronitens and uniformis, which they found to be
sisters, as in Ohlson et al.
Support for almost all nodes is very
strong. Their results add further
support to changes previously adopted by SACC in terms of breaking up Pipra into several genera.
To reconcile classification with their
phylogeny, they recommended the following changes at the genus level: (1) Xenopipo reduced to type species atronitens and its sister species X. uniformis; (2) Chloropipo is resurrected for the two Andean species, flavicapilla and unicolor; and (3) a new genus is described for holochlora, which is sister to Lepidothrix
and thus not closely related to Xenopipo
or Chloropipo.
This proposal is in three parts:
A.
Recognize Cryptopipo. I recommend a YES on this. The genetic data show clearly that holochlora is more closely related to Lepidothrix than to Xenopipo or Chloropipo,
and to include it within Lepidothrix
is the only other option given the data.
That option was considered and rejected by Ohlson et al. because: “it differs in
so many aspects of morphology and behavior that we are reluctant to include it
in that genus”. I agree
with that statement. A NO vote would
thus favor either inclusion of holochlora in Lepidothrix or retention of broadly defined Xenopipo.
B1.
Revise linear sequence of genera.
Our
current sequence is:
Neopelma
Tyranneutes
Ilicura
Masius
Corapipo
Antilophia
Chiroxiphia
Xenopipo
Machaeropterus
Dixiphia
Ceratopipra
Manacus
Heterocercus
Pipra
Lepidothrix
To alter the sequence to conform to the
Ohlson et al. tree, i.e., the addition of Chloropipo
and Cryptopipo, the following changes are needed in red:
Neopelma
Tyranneutes
Chloropipo
Ilicura
Masius
Corapipo
Antilophia
Chiroxiphia
Xenopipo
Machaeropterus
Dixiphia
Ceratopipra
Manacus
Heterocercus
Pipra
Cryptopipo
Lepidothrix
B2.
Further revise linear sequence of genera.
However, some additional changes are
advisable if we accept the Ohlson et al. tree as the best available data and
use on of the conventions for sequencing genera, i.e., for sister taxa, least-diverse
group first. For example, within the
Piprini in the figure, support is strong for Cryptopipo + Lepidothrix
as sister to the more diverse lineage (Heterocercus
+ Manacus + Pipra + Machaeropterus + Dixiphia + Ceratopipra). Within the
latter group, support varies from so-so to strong for the following
relationship: Heterocercus (Manacus + ((Pipra + (((Machaeropterus
+ ((((Dixiphia + Ceratopipra)))))))))). I
have mixed feelings on whether an overhaul of the sequence is warranted. On the other hand, these are the best
available data, and our previous rearrangement of the sequence hardly leaves
our current one with much of an historical legacy. Therefore, I lean towards going all the way
on the rearrangement at this point – as long as we’re going to make some
changes, might as well make all those indicated by the data, i.e.:
Tyranneutes
Neopelma
Chloropipo
Antilophia
Chiroxiphia
Ilicura
Masius
Corapipo
Xenopipo
Cryptopipo
Lepidothrix
Heterocercus
Manacus
Pipra
Machaeropterus
Dixiphia
Ceratopipra
This is exactly the sequence
recommended by Ohlson et al. (2013) except for the flip-flop of the groups (Antilophia + Chiroxiphia) and (Ilicura
+ ((Masius + Corapipo))).
C.
Add subfamilies. Ohlson et al. (2013) found the same pattern
as Tello et al. (2009) and McKay et al. (2010), namely a deep division within
the family, with Tyranneutes and Neopelma forming one lineage, and the
rest of the genera in the other. I
support recognition of this major division with subfamily rank for the two
lineages.
Literature
Cited:
McKAY, B. D., F. K.
BARKER, H. L. MAYS JR., S. M. DOUCET, AND G. E. HILL. 2010.
A molecular phylogenetic hypothesis for the manakins (Aves: Pipridae). Molecular Phylogenetics and Evolution 55:
733-737.
OHLSON, J., J. FJELDSÅ, AND P. G. P
ERICSON. 2013. Molecular phylogeny of the manakins (Aves: Passeriformes: Pipridae),
with a new classification and the description of a new genus. Molecular Phylogenetics and Evolution 69:
796–804.
TELLO, J. G., MOYLE, R.
G., D. J. MARCHESE, AND J. CRACRAFT.
2009. Phylogeny and phylogenetic classification of the tyrant flycatchers, cotingas, manakins, and their allies (Aves:
Tyrannides). Cladistics 25: 1-39.
Van Remsen, October
2013
__________________________________________________________________________________________________________________
Comments
from Stiles: “YES. The
Ohlson et al. paper provides the most thorough and comprehensive genetic data
set so far, and makes the description of Cryptopipra
for holochlora necessary, and
recognition of two subfamilies desirable.
Their data do make me wonder if Corapipo
and Masius ought to be merged? It looks like Masius makes Corapipo
paraphyletic?”
Comments
solicited from Jan Ohlson:
“"I
thank Van Remsen for so quickly bringing our proposals for changes of Pipridae
classification to the SACC board. As might be expected, I fully endorse the
proposals put forward by Van, and I am also strongly in favor of the
alternative B2 regarding sequence of genera, as this best reflects the
phylogenetic tree given the conventions accepted by the SACC".
Comments
from Pacheco: “YES.
Eu concordo com as recomendações –
incluindo as adaptações propostas por Remsen – derivadas de dois amplos estudos.”
Comments from Zimmer: “YES on parts A, B
and C, based largely on the data of Ohlson et al. (2013).”
Comments
from Pérez-Emán: “YES
to A and B2. The new phylogenetic study on Pipridae by Ohlson et al. (2013)
found a paraphyletic Chloropipo (or Xenopipo) clearly requiring a
new generic name for C. holochlora (Cryptopipo). This study also
provides the basis for updating linear sequence in the family, considering the
most comprehensive phylogenetic hypothesis currently known. I would vote NO for
C, as I consider this taxonomic level should consider not only molecular data
but also other characters that unambiguously show support for these taxonomic
categories.”
Comments
from Cadena: “591A. YES.
Regarding Gary's comment on eventually lumping Corapipo and Masius, we
should note that the relevant node is rather poorly supported (only 0.74
posterior probability and 48% bootstrap). This lack of resolution and the
plumage differences between genera suggests we should be conservative for now.
“591C. NO. Not
because I think the phylogenetic split is not strong, but rather based on a
more philosophical point of view. As I have commented on other proposals
before, I think we need to be consistent in our classification across the
board. If we are going to recognize subfamilies (i.e. well supported clades
within families), then we need to do this across all families, not haphazardly
in those families for which someone happens to present a proposal. Do we want
to do this for all families? This would be a lot of work, and I don't think we
should. If we do, then where do we stop once we are done? If we recognize
subfamilies, then why not tribes, suborders, etc..? I say we should stick to
major taxonomic ranks; sure, they are arbitrary, but they are arguably more
manageable in terms of number and in the degree of consistency that our
taxonomy already has (e.g., we recognize families, but not subfamilies, in
every order).”