Proposal
(735) to South American Classification Committee
Modify linear sequences to reflect new
phylogenetic data
A. Placement of Anthocephala
in Trochilidae
B. Sequence
of families in Suliformes
C.
Sequence of species and genera in Cathartidae
D.
Sequence of genera in Rallidae
E.
Sequence of species in Fulica
F.
Sequence of species in Charadrius
G. Invert
Laridae and Rynchopidae
H.
Sequence of species in Megascops
I.
Sequence of families in Coraciiformes
J.
Sequence of species in Chloroceryle
K.
Sequence of genera in Picidae
L.
Sequence of species in Forpus
Under the umbrella of this proposal I
collected a batch of minor linear sequence changes that need to be implemented
to reflect new phylogenetic data. This
is just drudgery bookkeeping that has to be done, but hopefully only once, and
so we won’t have to fiddle with it again (the Rallidae a likely exception). I don’t like the instability, but if we have
the rules, we have to follow them. I
restricted this proposal to nonpasserines just to cut it down in size. I did not present much in the way of methods
in each case; these can be found in the cited papers if interested. In each case, the sequences follows the
standard conventions of listing first the branch with the fewest species, and
in the case of sisters, the northwestern-most taxon is listed first.
I
recommend a YES on all of them (unless of course someone finds a mistake),
although I’m not 100% certain in the case of the Chloroceryle proposal.
A.
Placement of Anthocephala in
Trochilidae
Our current linear sequence of genera
in the Trochilidae was based on McGuire et al. (2009), which did not have
samples of Anthocephala. Thus, we left this genus next to Adelomyia, following Schuchmann’s
HBW classification. McGuire et al.
(2014) were able to obtain two samples of Anthocephala,
and found that this taxon is distantly related to Adelomyia and is sister to Stephanoxis,
with strong support, and is a member of a small group of hummingbird genera
that seem to have little in common, especially biogeography:

Therefore,
the two species of Anthocephala
should be moved in the sequence to precede the two species of Stephanoxis (in sister groups with same
species richness, northwestern-most taxon is first).
B. Sequence of families in Suliformes
Our current sequence of families in the
Suliformes is Fregatidae, Sulidae, Phalacrocoracidae, and Anhingidae. A minor correction needs to be made to follow
standard conventions for sequencing. All
recent data point to a sister relationship between Phalacrocoracidae and
Anhingidae, e.g., here’s the figure from Prum et al. (2015):

Anhingidae having fewer species means
that it should precede its sister taxon Phalacrocoracidae. Pretty exciting stuff, I know.
C. Sequence of species and genera in
Cathartidae
Our current linear sequence is as
follows:
Cathartes aura Turkey Vulture
Cathartes burrovianus Lesser Yellow-headed Vulture
Cathartes melambrotus Greater Yellow-headed Vulture
Coragyps atratus Black Vulture
Sarcoramphus papa King Vulture
Vultur
gryphus Andean Condor
Johnson
et al. (2016; MPE 105: 193-199) published
a phylogeny of the Cathartidae with the following tree:
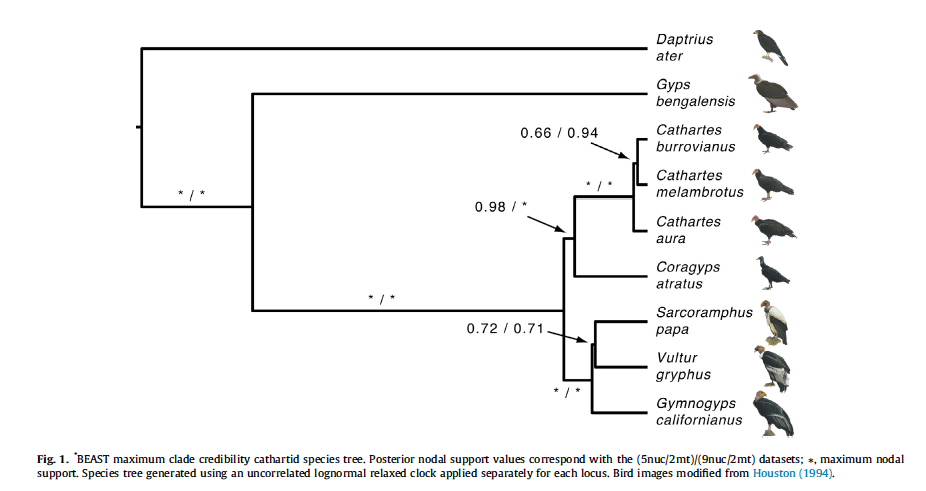
Converting this to a linear sequence
produces the following (treating Cathartes
as a polytomy):
Sarcoramphus papa King Vulture
Vultur
gryphus Andean Condor
Coragyps atratus Black Vulture
Cathartes aura Turkey Vulture
Cathartes burrovianus Lesser Yellow-headed Vulture
Cathartes melambrotus Greater Yellow-headed Vulture
D. Sequence of genera in Rallidae
Our current sequence of genera, based
mostly on historical momentum, is as follows:
RALLIDAE
(RAILS)
Coturnicops
Micropygia
Rallus
Aramides
Amaurolimnas
Anurolimnas
Laterallus
Crex
Porzana
Porphyriops
Mustelirallus
Pardirallus
Gallinula
Porphyrio
Fulica
García-R et al. (2014; MPE 81: 96-108) published a phylogeny
that lacks a lot of oddball genera, as you can imagine from such a globally
distributed family, but included most genera in the SACC area. The tree is too big to screen-grab (let me
know if you need the pdf; also, it is a little confusing because of taxonomic
differences). The big differences from
traditional relationships are that Porphyrio
is not closely related to Gallinula, and
that true Porzana are in the same
lineage Gallinula and Fulica.
They found strong support for the following relationships (extralimital
taxa pruned), with indentations used to indicate relationships:
Crex
Rallus
Porphyrio
Anurolimnas
Laterallus
Coturnicops
Mustelirallus
Pardirallus
Amaurolimnas
Aramides
Porphyriops
Porzana
Gallinula
Fulica
They did not have a sample of Micropygia, so we could keep it next to Coturnicops
to reflect reflect traditional ideas on its relationship. Because of the polyphyly of genera such as Laterallus, Porzana, and even Gallinula,
and the limited number of species sampled, I suspect we may have to do more
fiddling with this sequence once the family is more broadly sampled. Nonetheless, the sequence above will be much
closer to the eventual final sequence than our current one.
E. Sequence of species in Fulica
Our current sequence in Fulica
is as follows:
Fulica armillata Red-gartered Coot
Fulica rufifrons Red-fronted Coot
Fulica gigantea Giant Coot
Fulica cornuta Horned Coot
Fulica americana American Coot
Fulica ardesiaca Slate-colored Coot
Fulica leucoptera White-winged Coot
García-R et al. (2014; MPE 81: 96-108) had good sampling within
Fulica, with strong support for much
of the topology – a screen shot of that section of their tree is below. Of interest is that F. rufifrons is sister to all other coots, and that South America,
by traditional biogeographic reasoning, is the center origin for coots.
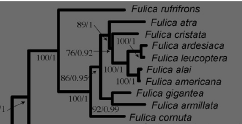
Translated this topology to a sequence produces the
following:
Fulica rufifrons Red-fronted Coot
Fulica cornuta Horned Coot
Fulica gigantea Giant Coot
Fulica armillata Red-gartered Coot
Fulica americana American Coot
Fulica ardesiaca Slate-colored Coot
Fulica leucoptera White-winged Coot
F. Sequence of species in Charadrius
Our current sequence in Charadrius is as follows:
Charadrius semipalmatus Semipalmated Plover
Charadrius melodus Piping Plover
Charadrius wilsonia Wilson's Plover
Charadrius vociferus Killdeer
Charadrius nivosus Snowy Plover
Charadrius collaris Collared Plover
Charadrius alticola Puna Plover
Charadrius falklandicus Two-banded Plover
Charadrius modestus Rufous-chested Dotterel
Joseph et al. (1999) and Dos Remedios
et al. (2015; MPE) produced
phylogenies that show that there are two major divisions within Charadrius, and that broadly defined Charadrius is paraphyletic with respect
to extralimital, Old World Eudromias. The tree from the latter is pasted in here:
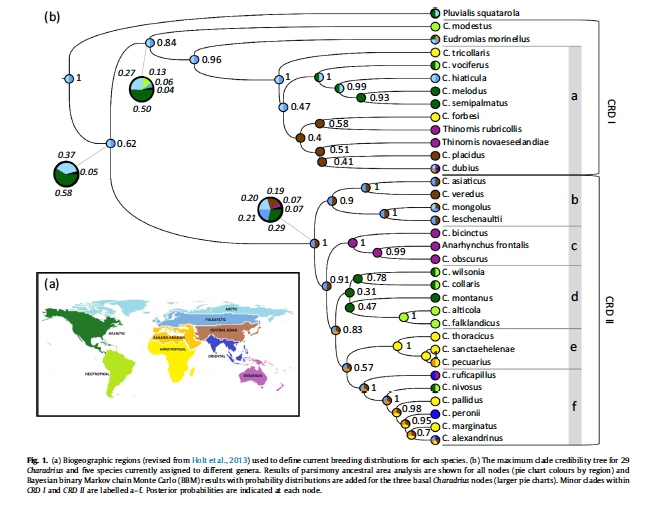
It looks to me that Zonibyx should be resurrected for modestus and that Charadrius should be split into two genera. Regardless, until such a reclassification is
proposed, converting this tree to a linear sequence for just our species
produces the following sequence, here with indentations and spacing to try to
capture the topology:
Charadrius modestus Rufous-chested Dotterel
Charadrius
vociferus
Killdeer
Charadrius
semipalmatus
Semipalmated Plover
Charadrius melodus Piping Plover
Charadrius
wilsonia
Wilson's Plover
Charadrius
collaris
Collared Plover
Charadrius
alticola
Puna Plover
Charadrius
falklandicus
Two-banded Plover
Charadrius
nivosus
Snowy Plover
G. Reverse Laridae and Rynchopidae
All data indicate that the Laridae and
Rynchopidae are sister families.
However, if linear sequences are to follow conventions, rather than
tradition, Rynchopidae should clearly precede the much more diverse group
Laridae, and the sequence of the two should be reversed.
H. Sequence of species in Megascops
Our current sequence in Megascops is as follows:
Megascops choliba Tropical Screech-Owl
Megascops roboratus Peruvian Screech-Owl
Megascops koepckeae Koepcke's Screech-Owl
Megascops clarkii Bare-shanked Screech-Owl
Megascops colombianus Colombian Screech-Owl
Megascops ingens Rufescent Screech-Owl
Megascops petersoni Cinnamon Screech-Owl
Megascops marshalli Cloud-forest Screech-Owl
Megascops watsonii Tawny-bellied Screech-Owl
Megascops guatemalae Vermiculated Screech-Owl
Megascops hoyi Montane Forest Screech-Owl
Megascops atricapilla Black-capped Screech-Owl
Megascops sanctaecatarinae Long-tufted Screech-Owl
Megascops albogularis White-throated Screech-Owl
Dantas et al. [including Daniel and
Mark] (2016; MPE 94: 626–634) produced a
comprehensive phylogeny of the genus, and their tree is below:
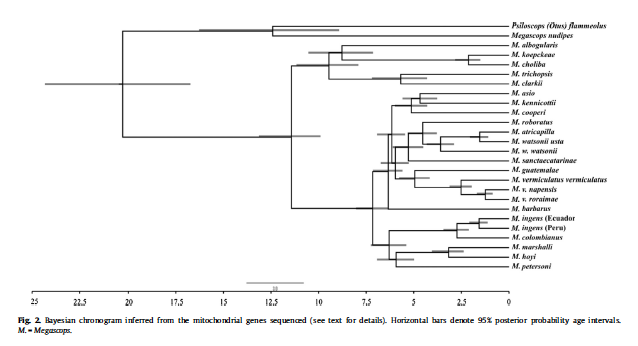
Converting this to a linear sequence,
with extralimital taxa not included, produces the following:
Megascops clarkii Bare-shanked Screech-Owl
Megascops albogularis White-throated Screech-Owl
Megascops choliba Tropical Screech-Owl
Megascops koepckeae Koepcke's Screech-Owl
Megascops colombianus Colombian Screech-Owl
Megascops ingens Rufescent Screech-Owl
Megascops petersoni Cinnamon Screech-Owl
Megascops marshalli Cloud-forest Screech-Owl
Megascops hoyi Montane Forest Screech-Owl
Megascops guatemalae Vermiculated Screech-Owl
Megascops sanctaecatarinae Long-tufted Screech-Owl
Megascops roboratus Peruvian Screech-Owl
Megascops watsonii Tawny-bellied Screech-Owl
Megascops atricapilla Black-capped Screech-Owl
I. Sequence of families in Coraciiformes
Our current sequence of families is
Alcedinidae and Momotidae. The rationale
behind this is that the relationship among the three families in the New World
was always assumed to be Alcedinidae + (Todidae + Momotidae). However, all recent data sets e.g. Prum et
al. (2015) show that the relationship is Todidae + (Alcedinidae + Momotidae):

Therefore, the sequence should be
Momotidae, then Alcedinidae (the more diverse family). Yawn.
J. Sequence of species in Chloroceryle
Our current sequence is as follows
Chloroceryle amazona Amazon Kingfisher
Chloroceryle americana Green Kingfisher
Chloroceryle inda Green-and-rufous Kingfisher
Chloroceryle aenea American
Pygmy Kingfisher
Moyle et al. (2006), however, found the
following relationships:
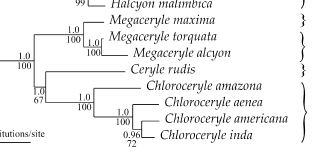
Thus, to make the sequence reflect
these data, C. aenea needs to be moved, as follows:
Chloroceryle amazona Amazon Kingfisher
Chloroceryle aenea American
Pygmy Kingfisher
Chloroceryle americana Green Kingfisher
Chloroceryle inda Green-and-rufous Kingfisher
Note that support for the americana-inda sister relationship is
not rock solid, and so that could be a reason to vote for stability until
support for that node solidifies.
K. Sequence of genera in Picidae
Our linear sequence of
genera in the Picidae is as follows:
Picumninae
Picumnus
Picinae
Melanerpes
Sphyrapicus
Picoides
Veniliornis
Piculus
Colaptes
Celeus
Dryocopus
Campephilus
Benz et al. [including Mark] (2006)
published a phylogeny of the Picidae that generated the following tree, which
is at odds with the sequence of genera in traditional classifications:
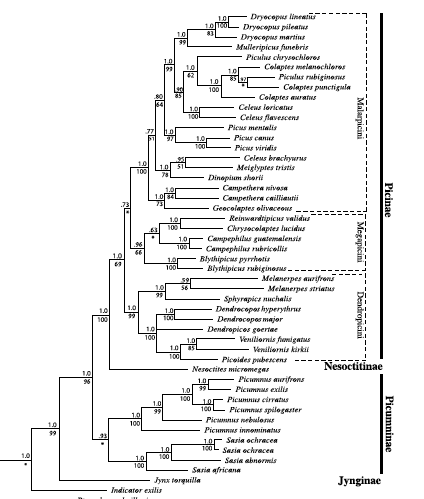
To make our sequence reflect these data
(accounting for species missing in the tree), the sequence needs to be as
follows:
Picumninae
Picumnus
Picinae
Sphyrapicus
Melanerpes
Picoides
Veniliornis
Campephilus
Dryocopus
Celeus
Piculus
Colaptes
Unfortunately, this still keeps Campephilus and Dryocopus adjacent, which
perpetuates the misconception that they are related, but that’s the way the
sequence works out.
L. Sequence of species in Forpus
Our current sequence in Forpus is as follows:
Forpus passerinus Green-rumped
Parrotlet
Forpus xanthopterygius Blue-winged
Parrotlet
Forpus conspicillatus Spectacled
Parrotlet
Forpus modestus Dusky-billed
Parrotlet
Forpus coelestis Pacific
Parrotlet
Forpus xanthops Yellow-faced
Parrotlet
Smith et al. (2013) found the following
relationships:
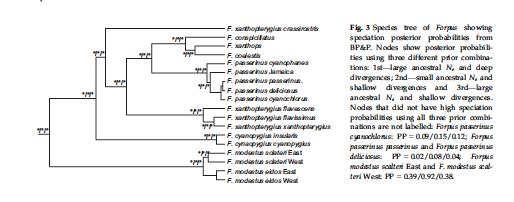
Converting this to a linear sequence
produces the following:
Forpus modestus Dusky-billed
Parrotlet
Forpus xanthopterygius Blue-winged
Parrotlet
Forpus passerinus Green-rumped
Parrotlet
Forpus conspicillatus Spectacled
Parrotlet
Forpus coelestis Pacific
Parrotlet
Forpus xanthops Yellow-faced
Parrotlet
___________________________________________________________
Comments from Stiles: “YES to all.
Necessary given the evidence now available, while recognizing that for some
families (e. g., Rallidae and Trochilidae), further changes might be necessary
as further evidence accrues.”
Comments from Areta: “YES to all changes. I have not found mistakes.”
Comments from Pacheco:
“YES to all updates. The small changes in the sequences are corroborated by the
most recent phylogenies.”
Comments from Zimmer: “YES to all. Given the most recent
data, and our conventions, this all seems
like the way to go.”
Comments from Jaramillo:
“E. YES: rufifrons is the oddball in
the field, if you ignore size and horns. It is much more Gallinule-like in
behavior, habitat choice, and even overall structure. I am not surprised on its
placement.”
Comments from Claramunt:
“Some relationships are not well resolved (low support)
and things may change in the near future but overall, the changes are an improvement.
All the changes seem correct according to our (new) sequencing criteria.”