Proposal (869) to South American Classification Committee
Revise linear
sequence of species of macaws
Description of the
problem:
This is a repurposed, minimally modified proposal
sister to that passed in NACC (Chesser et al. 2020; 2020-B-6) for resequencing Ara macaws.
Although the Cuban Macaw Ara tricolor has
been extinct since the mid-19th century, it is the best-known of the
several putative Antillean endemic macaws, all extinct and several of dubious
validity (Wiley and Kirwan 2013). Based on plumage, Ara tricolor has
been suggested to be closely related to the other but much larger (Forshaw and
Cooper 1973) red macaws, A. macao and A. chloropterus. Unlike
other Antillean macaws, there are several extant specimens of A. tricolor,
19 skins and fragments from three fossil sites (Olson and Suárez 2008). The
skins have not previously been sampled for genetic analysis.
New information:
Two A. tricolor specimens are held at the
Swedish Museum of Natural History, and toepads of both were sampled (Johansson
et al. 2018). Complete mtDNA genomes were sequenced for five of the seven
species of macaws that occur in the NACC area, all except A. ambiguus
and A. chloropterus, for which partial mitochondrial sequence (16S and
CO1) was obtained. Contrary to expectations that it would prove most closely
related to the two extant red macaws, in this phylogeny A. tricolor is
sister to the clade that includes both large red and large green macaws (A.
militaris and A. macao).
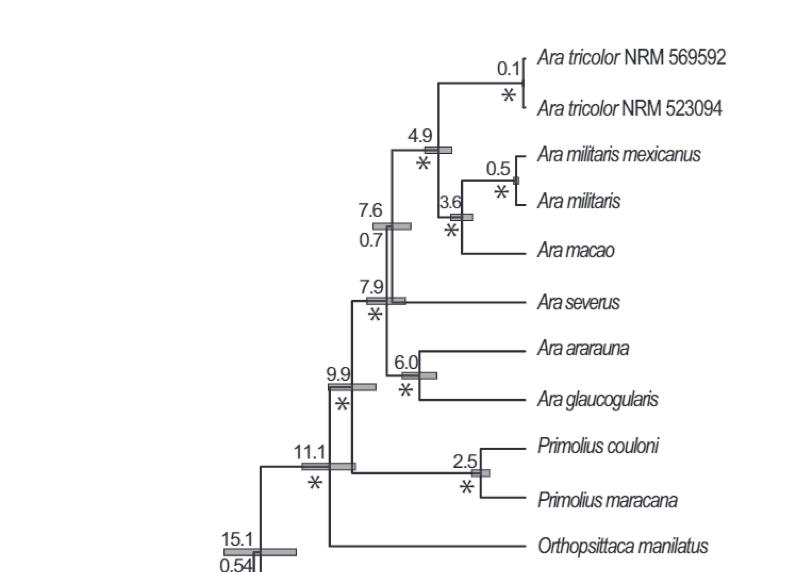
Relevant
portion of Figure 1 in Johansson et al. (2018), a phylogeny based on complete
mitochondrial sequences. Estimated divergence dates are above the nodes and
posterior probabilities below (* = 1.0 pp).
Johansson
et al. (2018) also produced a phylogeny based on the partial mitochondrial
data; this included all species in our area. The top half of this phylogeny is
poorly supported, but the clade containing tricolor
is relatively robust and supports the position of tricolor as sister to a clade containing militaris and macao, as well as the two species not sampled
in the mt-genome phylogeny, ambiguus and
chloropterus. This phylogeny
indicates that ambiguus is sister to
and very closely related to militaris,
and that chloropterus is also part of
this clade, but that its placement is unresolved relative to macao and militaris/ambiguus. Ara
rubrogenys is poorly supported as sister to Ara severus.
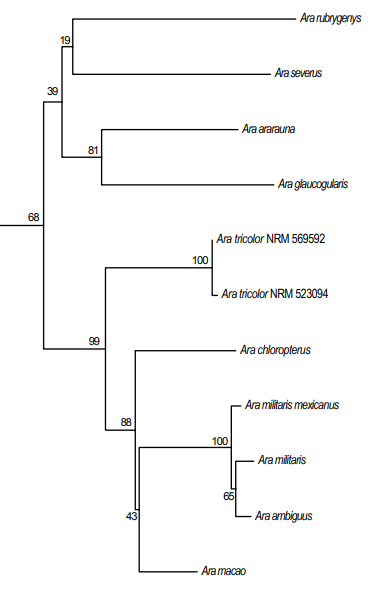
Relevant
portion of Figure S1 of Johansson et al. (2018), a phylogeny based on sequences
of the mitochondrial genes 16S and CO1.
With acceptance of this proposal, these macaws have
now been resequence as follows for the NACC area:
Ara ararauna
Ara severus
Ara tricolor
Ara militaris
Ara ambiguus
Ara macao
Ara chloropterus
In
generating this linear sequence, we considered macao + chloropterus + militaris/ambiguus to form a 3-way polytomy, and placed militaris/ambiguus last
because it consists of more species, and macao
first because of its more northerly distribution than chloropterus.
The SACC area Ara
macaws (Remsen et al. 2020) are currently sequenced as follows:
Ara ararauna
Ara glaucogularis
Ara militaris
Ara ambiguus
Ara macao
Ara chloropterus
Ara rubrogenys
Ara severus
Following SACC conventions and for congruence with
NACC, the sequence should be:
Ara ararauna
Ara glaucogularis
Ara severus
Ara rubrogenys
Ara militaris
Ara ambiguus
Ara macao
Ara chloropterus
Given that the remainder of the Johansson et al.
(2018) phylogeny is not densely sampled, this proposal focused only on macaws.
Recommendation:
We
recommend adopting these minor sequence changes.
Literature cited:
Chesser, R.T., S.M. Billerman, K.J. Burns, C. Cicero, J.L. Dunn, A.W.
Kratter, I.J. Lovette, N.A. Mason, P.C. Rasmussen, J.V. Remsen, Jr., D.F.
Stotz, and K. Winker (2020). Sixty-first Supplement to the American
Ornithological Society’s Check-list of North American Birds. The Auk:
Ornithological Advances. https://doi.org/10.1093/auk/ukaa030.
Forshaw, J. & W. Cooper. 1973. Parrots of the
world. Doubleday & Co., Garden City, New York.
Johansson, U.S., P.G.P. Ericson, M.P.K. Blom &
M. Irestedt. 2018. The phylogenetic position of the extinct Cuban Macaw Ara
tricolor based on complete mitochondrial genome sequences. Ibis 160:
666-672.
Olson, S.L. & W. Suárez. 2008. A fossil cranium
of the Cuban Macaw Ara tricolor (Aves: Psittacidae) from Villa Clara
Province, Cuba. Caribbean Journal of Science 44: 287-290.
Wiley, J.W. & G.M. Kirwan. 2013. The extinct
macaws of the West Indies, with special reference to Cuban Macaw Ara
tricolor. Bulletin of the British Ornithologists’ Club 133: 125-156.
Pamela C. Rasmussen,
July 2020.
Comments
from Remsen:
“YES. Required book-keeping to follow
the conventions of linear sequencing in a phylogenetic classification. The genetic data aren’t great – two mtDNA
loci – but at least they represent a phylogenetic hypothesis to be tested and
modified as needed … in contrast to the current sequence, which in Ara
based is based on historical momentum without a explicit rationale (as far as I
know).”
Comments
from Robbins:
“YES. I vote yes for the sequence. The analyses by Provost et al. (2017) provide
stronger support for some of these nodes. If one looks at the Johansson et al. paper, it
fails to cite Provost et al. Take a look
at the pertinent figure in the reference:
“Resolving a phylogenetic hypothesis for parrots: implications from systematics
to conservation Kaiya L. Provost, Leo Joseph & Brian Tilston
Smith Published online: 01 Nov 2017.
Austral Ornithology.”
Comments
from Stiles:
“YES. The new sequence looks OK to me.”
Comments from Jaramillo: “YES. This is congruent with
multiple published data including Provost et al. 2017 which was not cited in
the proposal.”
Comments
from Areta:
“YES. The different phylogenies coincide, so we should re-sort species to
follow them as developed by Pam.”
Comments from Pacheco: “YES. Consistent with the latest
results.”
Comments from Claramunt: “YES. The proposed sequence
follows SACC conventions. I first thought that ambiguus+militaris
should be listed last as it is the most diverse clade in the 3-way polytomy but
I see that is not what our criteria state.
Comments from Bonaccorso: “YES. The sequence is consistent
with the evidence at hand.”