Proposal (883) to South
American Classification Committee
Recognize
sixteen species in the Grallaria rufula complex
Background. Hellmayr (in Cory and Hellmayr 1924), having synonymized saturata Domaniewski & Stolzmann,
1918, with the nominate form, recognized four subspecies of Grallaria rufula: rufula Lafresnaye, 1843; obscura Berlepsch and Stolzmann, 1896; spatiator Bangs, 1898; and
occabambae Chapman, 1923. Three additional subspecies (cajamarcae Chapman,
1927; cochabambae Bond and Meyer de Schauensee, 1940; and saltuensis Wetmore,
1946) were later described, some of them, especially saltuensis,
characterized by rather distinctive plumage. Thus, seven subspecies of G. rufula were generally recognized
(Peters 1951) when Graves described Grallaria
blakei (1987). Despite its similarity to G. rufula, Graves documented that G. blakei was specifically distinct based on elevational parapatry
(possibly sympatry) as well as plumage and vocal characters.
The
discovery of two species of the G. rufula
complex in close proximity raised the possibility that other populations in the
complex were also specifically distinct. Graves studied the morphology of the
group in the 1990s, measuring coloration with a spectrophotometer, but he did
not come to taxonomic conclusions. Identification of plumage differences among
populations is made difficult by individual plumage variation, as noted by
Hellmayr (op. cit.).
In
collaboration with Graves, M. and P. Isler began analyzing vocal recordings in
the 1990s. As the years went by, ornithological expeditions to the Andes
produced new information regarding the complex, including range extensions,
specimens, and vocal recordings. Notably, populations of G. rufula and presumed G. blakei
were found in mountain ranges in which they were previously unknown. In
comments deposited in recording archives, recordists noted differences in
vocalizations among populations, and the need for a thorough analysis of the
complex was noted in the ornithological literature (e.g., Schulenberg et al.
2007, Greeney 2018).
In
response, T. Chesser and M. Isler undertook a comprehensive study of the
systematics and evolution of the Grallaria
rufula complex focusing primarily on genetics and vocalizations. Other
individuals became deeply involved as co-authors, and many others contributed
vital material or analytical support as the studies continued (see
Acknowledgments in Chesser et al. 2020 and Isler et al. 2020).
Analysis and Results. We sequenced
nuclear and mitochondrial DNA for 80 individuals from across the distribution
of the G. rufula complex to determine the extent of genetic variation
between and within populations (Chesser et al. 2020) Our results revealed 18
geographically coherent clades separated by substantial genetic divergence: 14
within rufula, three within blakei, and one corresponding to G. rufocinerea (Bicolored Antpitta), a
species with distinctive plumage found to be nested within the complex (Figs.
1-3). Working within the framework of the molecular phylogeny, we
reexamined species limits in the G. rufula complex, basing taxonomic
recommendations on diagnostic differences in vocalizations and considering
identifiable differences in plumage where pertinent (Isler et al. 2020). Owing
to the large number of populations involved, analyses were initially confined
to populations within eight principal clades that have accrued substantial
genetic divergence (> 5% uncorrected pairwise distance in the mitochondrial
gene ND2) and then extended to comparisons between principal clades. Vocal
differences between study populations had to be discrete, typically
non-overlapping character states that have the potential for unambiguous signal
recognition. Following the three character guideline developed empirically for
the closely related Thamnophilidae (Isler et al. 1998), we identified 16
populations for species designation, including seven populations previously
described as subspecies (including saturata) and six new species. We identified less robust vocal differences between
populations within the newly recognized species G. occabambae that were
designated as subspecies .
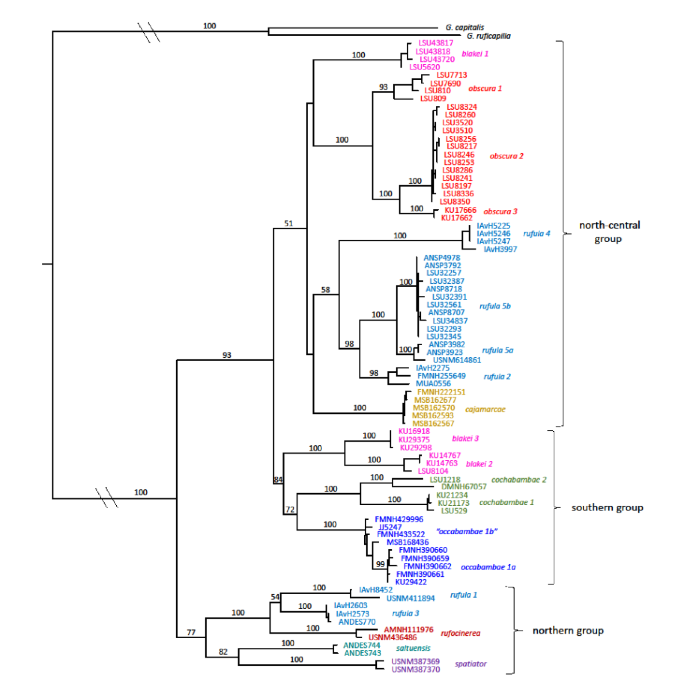
Figure 1. Most likely tree for
the Grallaria rufula complex, based on the combined mitochondrial and
nuclear data, produced using the program RAxML. Numbers above nodes represent
bootstrap support values based on 100 bootstrap replicates. This tree differs
from the ML bootstrap tree largely in its placement of rufula 4; hence,
the low support values in this part of the tree.
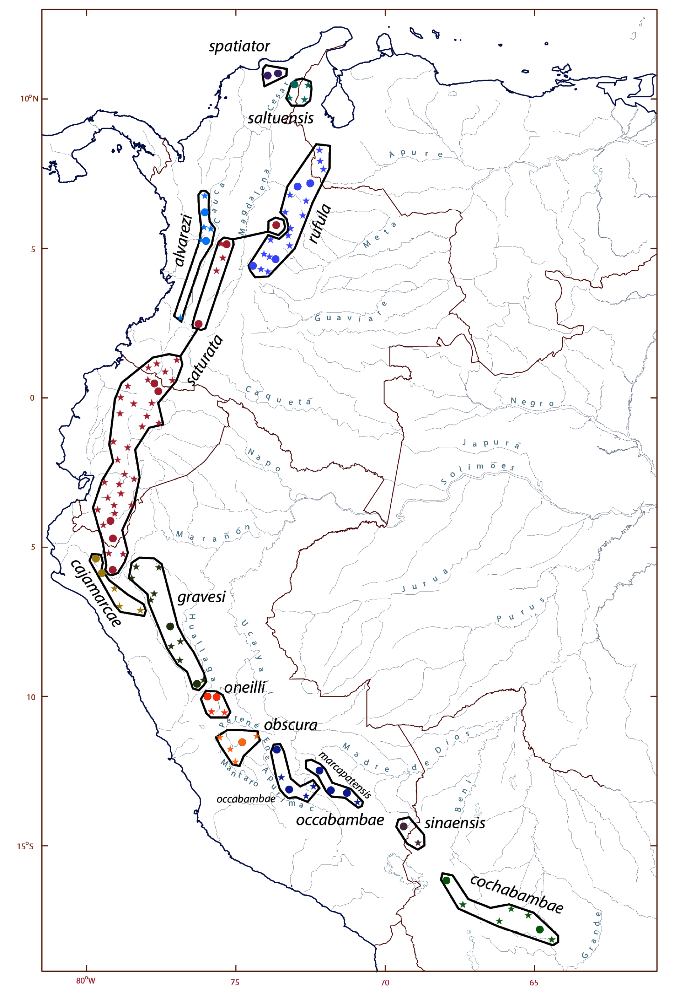
Figure 2. Map showing the
taxonomy and distribution of the Grallaria rufula complex as revised.
Part 1: populations formerly ascribed to G. rufula.
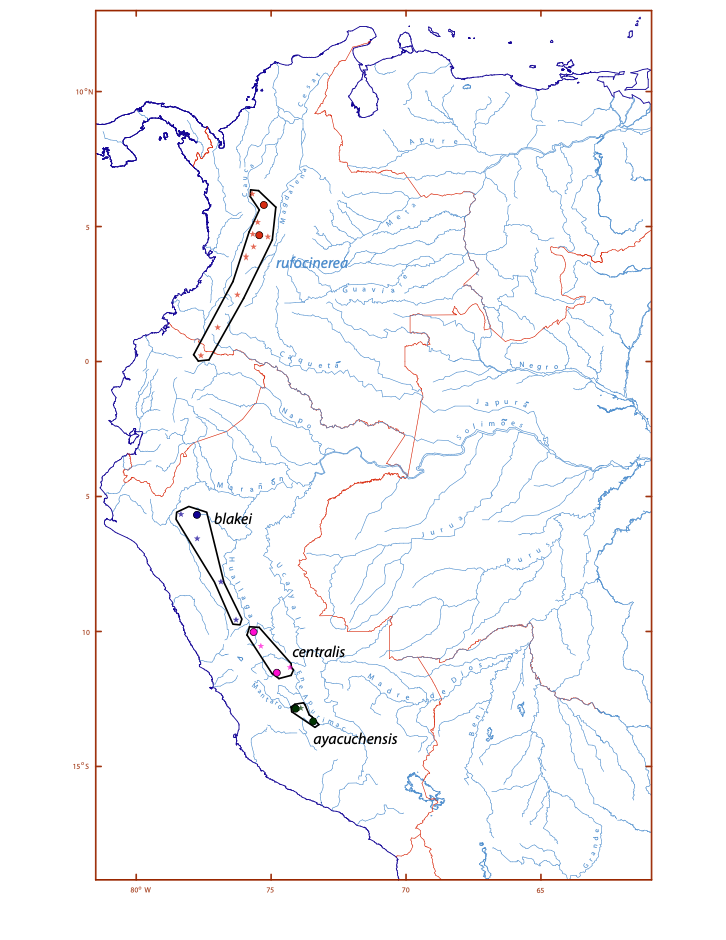
Figure 3. Map showing the
taxonomy and distribution of the Grallaria rufula complex as revised.
Part 2: populations formerly ascribed to G. blakei and G. rufocinerea.
Results are not summarized here because of the large number of taxa
involved, and reviewers are directed to the text and Appendix 1 of Isler et al.
(2020).
Recommendation. We recommend that the following sixteen taxa be recognized as species.
Thirteen of these would be newly recognized species, whereas three, marked by
asterisks, are currently considered species.
Principal
Clade A
1.
Grallaria saltuensis Wetmore, 1946.
Perijá Antpitta. Subspecies elevated to species. Includes molecular clade saltuensis.
2.
Grallaria spatiator Bangs, 1898.
Sierra Nevada Antpitta. Subspecies elevated to species. Includes molecular
clade spatiator.
Principal
Clade B
3.
*Grallaria rufula Lafresnaye, 1843.
Muisca Antpitta. Includes molecular clades rufula 1 and rufula 3.
4.
*Grallaria rufocinerea Sclater &
Salvin, 1879. Bicolored Antpitta. Includes molecular clade rufocinerea.
Principal
Clade C
5.
Grallaria alvarezi Cuervo, Cadena,
Isler, & Chesser. Chami Antpitta. New species. Includes molecular clade rufula
4.
6.
Grallaria saturata Domaniewski & Stolzmann, 1918. Equatorial Antpitta.
Subspecies resurrected and elevated to species. Includes molecular clades rufula
2 and rufula 5a-5b.
Principal
Clade D
7. Grallaria cajamarcae (Chapman, 1927). Cajamarca Antpitta. Subspecies elevated to species. Includes molecular clade
cajamarcae.
Principal
Clade E
8. *Grallaria blakei Graves, 1987. Chestnut Antpitta. Includes
molecular clade blakei 1.
Principal
Clade F
9.
Grallaria gravesi Isler, Chesser,
Robbins & Hosner. Graves’s Antpitta. New species. Includes molecular clade obscura
1.
10.
Grallaria oneilli Chesser &
Isler. O'Neill’s Antpitta. New species. Includes molecular clade obscura 2.
11. Grallaria
obscura Berlepsch & Stolzmann, 1896. Junín Antpitta. Subspecies
elevated to species. Includes molecular clade obscura 3.
Principal
Clade G
12.
Grallaria centralis Hosner, Robbins,
Isler, & Chesser. Oxapampa Antpitta. New species. Includes molecular clade blakei
2.
13.
Grallaria ayacuchensis Hosner,
Robbins, Isler, & Chesser. Ayacucho Antpitta. New species. Includes
molecular clade blakei 3.
Principal
Clade H
14. Grallaria
occabambae (Chapman, 1923). Urubamba Antpitta. Subspecies elevated to
species. Includes subspecies occabambae and marcapatensis, Isler
& Chesser. Includes molecular clade occabambae
1a-1b.
15. Grallaria sinaensis Robbins, Isler, Chesser, & Tobias. Puno
Antpitta. New species. Includes molecular clade cochabambae 1.
16. Grallaria
cochabambae Bond & Meyer de Schauensee, 1940. Bolivian Antpitta.
Subspecies elevated to species. Includes molecular clade cochabambae 2.
References.
Chesser,
R.T., Isler, M.L., Cuervo, A.M., Cadena, C.D., Galen, S.C., Bergner, L.M.,
Fleischer, R.C., Bravo, G.A., Lane, D.F., & Hosner, P.A. (2020)
Conservative plumage masks extraordinary phylogenetic diversity in the Grallaria rufula (Rufous Antpitta) complex of the humid
Andes. The Auk: Ornithological Advances
137:1–25.
Cory,
C.B. & Hellmayr, C.E. (1924) Catalogue of birds of the Americas and the
adjacent islands. Pteroptochidae - Conopophagidae - Formicariidae. Field Museum of Natural History Zoological
Series, no. 13, part 3, 1–369.
Graves,
G.R. (1987) A cryptic new species of antpitta (Formicariidae: Grallaria) from the Peruvian Andes. Wilson Bulletin, 99, 313–321.
Greeney,
H.F. (2018) Antpittas and Gnateaters.
Helm Identification Guides. Helm,
London & New York, 496 pp.
Isler,
M. L., Chesser, R.T., Robbins, M.B., Cuervo, A.M., Cadena, C.D., & Hosner,
P.A. (2020) Taxonomic evaluation of the Grallaria
rufula (Rufous Antpitta) complex (Aves: Passeriformes: Grallariidae)
distinguishes sixteen species. Zootaxa 4817:1–74.
Isler,
M.L., Isler, P.R. & Whitney, B.M. (1998) Use of vocalizations to establish
species limits in antbirds (Passeriformes: Thamnophilidae). The Auk, 115, 577–590.
Peters,
J.L. (1951) Check-list of Birds of the World, vol. 7. Museum of Comparative
Zoology, Cambridge, Massachusetts, 318 pp.
Schulenberg,
T.S., Stotz, D.F., Lane, D.F., O'Neill, J.P. & Parker III, T.A. (2007) Birds of Peru. Princeton University
Press, Princeton, New Jersey, 656 pp.
Morton
L. Isler, September 2020
Comments
from Areta:
“YES to all splits, descriptions and English names.
It is amazing that some populations (notably rufula 1 and rufula 2)
may still be afforded species status in the future when more vocalizations and
further evidence is obtained.”
Comments
from Robbins:
“YES, to recognizing all of these cryptic Grallaria
as species.”
Comments from Zimmer: YES to recognizing all splits, new taxa
descriptions, and suggested English names as detailed in the Isler et al. (2020)
and as summarized in the Proposal. The
authors of this monumental study (literally decades in the making) are to be
commended for the thoroughness with which they have unraveled this complicated
taxonomic puzzle.”
Comments from Stiles: “YES to all
(recognizing the 16 species). Because the E-names have already been published
in a companion paper, I see no reason not to accept these as published also.”
Comments from Claramunt: “This is a fantastic study
and a real eye-opener regarding species limits in grallariids.
Overall, the study is solid and the evidence overwhelming in most cases.
However, I am hesitant to give the proposal an overall YES. I wanted to comment
and hear opinions on a couple of cases. The congruence of diagnostic traits
across datasets, and in particular the congruence between the mitochondrial and
nuclear trees, clearly indicate the existence of 13 lineages here. Evidence for
further subdivisions is no so strong and clear. Given that the sample sizes and
geographical coverage of some of the (now narrowly endemic) lineages was low,
there is room for being conservative in a couple of cases. In addition, if I
have to give an opinion from the perspective of the biological species concept,
the evidence is tenuous for many lineages. The evidence for intrinsic
reproductive isolation is very indirect. In particular, there are several pairs
of lineages that have very similar songs, and I don’t think that the
diagnosability criteria imported from a different family (Thamnophilidae)
mostly from a different biome (the Amazon) can be used to infer song-based
reproductive isolation in Grallaria. The cases that I consider don’t
have strong evidence for multiple species are:
“Clade F. Songs sound similar within this clade and molecular data
is not conclusive. Song differences seem to be diagnostic among the three taxa
but songs still sound similar, so the question is whether those subtle
differences are enough to maintain reproductive isolation in sympatry. In
addition, the nuclear data did no sort these three taxa and the molecular
species delimitation indicated 2, not three species. So, I think we can elevate
obscura to the species rank but include oneilli and gravesi
as subspecies.
“Clade G. (ayacuchensis and centralis) The full
congruence between nuclear and mitochondrial data suggests two separate
lineages. But songs are very similar and the song samples of ayacuchensis
are from one extreme of its distribution. It remains to be demonstrated that
the subtle song differences are sufficient as reproductive barriers. So,
definitely there are two independent lineages here, but who knows if they have
intrinsic reproductive barriers.
“Clade H. I think we should play conservative in this case and
recognize only two species (cochabambae and ocabambae).
not three. The nuclear data supports the separation between the cochabambae
and occabambae groups (barely) but sinaensis (cochabambae1) is
nested within cochabambae. Songs
seem similar. and we don’t know if the differences are sufficient for
reproductive isolation. The sampling is
somewhat sparse, maybe reflecting actual discontinuities but chances are that
there are intervening populations that can be studied and be informative.”
Comments
from Jaramillo:
“YES – And may I add how impressive this work is, unbelievable. To think that
there are people out there that think that all discoveries have already been
made. This confirms that in some cases, we are at the beginning of
understanding, not far along on the journey! Commendable.”
Comments
from Lane:
“YES to all. I was hesitant with the very same cases
outlined by Santiago, but after listening to recordings just now, I think I can
understand the reasoning used to propose those taxa as species rather than
subspecies, namely: either the long song is distinctive (more often than not),
or characters of the short song are measurably distinct (less important, particularly
in the case of the two taxa proposed to fall within G. occabambae).
So, even though these doubts caused me to hesitate in accepting the full slate
of species-level taxa, after listening to recordings, I think my doubts are
assuaged and I am willing to accept the results of this pair of papers as they
were presented.”
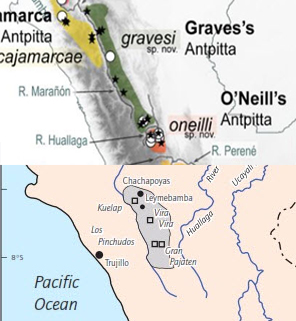
“Now, one thing I wonder: why not take a page from the Colombian
playbook, and give the English name "Chachapoyas Antpitta" to G.
gravesi? This civilization has been overshadowed by the Inca but it is
nevertheless one of the most important in Peru and South America. As such, it
seems that the broad overlap in the distribution of the antpitta and the
civilization (see map; see http://anthropology.iresearchnet.com/chachapoya-indians/)
would make this species an excellent choice to take on the moniker.
Response
to Claramunt’s comments from Mort Isler and Pete
Hosner”
“The
authors appreciate the care that Santiago Claramunt took in reviewing the rufula
species recommendations and his kind words regarding the papers. Before
discussing his clades of concern, a brief response to more general comments is
in order.
“The use of
the results from studies of the Thamnophilidae and its application to the
Grallariidae must be clarified. The thamnophilid studies provided an analytic
approach, not “diagnosability criteria” which has been erroneously taken to
mean a required number of diagnosable vocal characters for species
designation. The 1998 paper cited
stressed that the number of diagnosable vocal characters should be considered
“a point of reference, not a requirement” in species designations in which
vocal characters should be considered along with morphological and other
characters, phylogenetic results, and biogeographic considerations. This is the
approach that is applied in the paper and in the responses to Claramunt’s concerns.
“A second
point of concern are sample limitations and geographic coverage. The sampling
gaps Claramunt identifies are fine scale, only some ~150 km, over a study area
of over 4,000 km (roughly following the contour of the Andes. In this context,
all sampling gaps are minor, and not a substantive criticism. Also, there is no
evidence of clinal variation in the broader group over 4,000 km, so why would
it be expected over ~150 km? In both
papers we recognized sampling limitations in a number of regions and implored
the undertaking of additional field work to address these limitations. We would
argue, however, that it is vital from conservation and analytic concerns to
make decisions now, especially because the conservation need is so obvious, and
that current knowledge is sufficient to make them. New knowledge can and will
bring amendments.
“Third,
Claramunt advocates that there is only data congruence supporting 13 lineages.
This is a mis-interpretation of our data, there is molecular/vocal congruence
supporting the 16 lineages that we recommend recognizing as species (including blakei
& rufocinerea). What is true is that the nuclear data on its own
only identifies 13 clades. But this is not a solid argument against recognition
of all 18 species. These nuclear introns are slowly evolving and the lineages
are slow to sort. There is no empirical expectation for species monophyly based
on a few of nuclear introns, the lineage sorting process takes millions of
years, depending on generation length, population size, and stochasticity. We
can see this lineage sorting process in our data. Even with just the three
introns on their own, which admittedly is not a lot of data, we see that obscura1
and obscura3 are nested clades within obscura2. Hence, the
nuclear data is perfectly congruent with the existence of 16 species in the
rufula complex, which are each further supported by strong mitochondrial and
vocal differences.
“Turning to
the specific comments, in writing the taxonomic paper we spent considerable
time weighing the species recommendations that Santiago flagged. In each case
we believe that the vocal differences are sufficient (contrary to Claramunt)
within the context of the analytic approach described above. Without repeating
material available in the papers, the following briefly summarizes
considerations for each.
“Clade F.
Of the three clades cited, Clade F was the site of the greatest struggle in our
species recognition. Relating the three populations, the vocal study found that
obscura was the most distinct and that oneilli and gravesi
the least; the analysis of plumage found that oneilli was the most
distinct and gravesi and obscura the least; and genetically, oneilli
and obscura were sister to the
exclusion of gravesi least. Also, Claramunt suggested that the bGMYC analysis supported two rather than three species in
this group, but that was using a conservative threshold. The most-likely result
recognized all three as species. Although inconsistent, the extent of
differences among these populations led us to conclude that the three populations
have evolved independently to the point that they should be considered species.
Considering them as species, rather than subspecies, makes the decision
consistent with the other species recommendations in the paper, in contrast
with the subspecies split in occabambae. Interestingly, their
biogeography indicates that gravesi is isolated from its neighbor, oneilli,
by the Rio Huallaga, a major river barrier, and oneilli and obscura
by lesser barriers.
“Clade G. Significant differences in
both long and short songs as well as plumage support the consideration of ayacuchensis
and centralis as distinct species. It is notable that the range of centralis
is restricted a narrow elevational zone (2400–2700 m.) in regions where oneilli
and obscura occupy higher elevations. In contrast, ayacuchensis
inhabits a wider swath of elevations (2500–3700 m.) and habitats, suggesting
the possibility of differences in their ecology.
“Clade H. Significant differences in
both long and short songs as well as plumage support the consideration of sinaensis
and cochabambae as distinct species. Field work is needed to define
their geographic relationship which conceivably might be parapatric. The
paraphyly in the nuclear tree appears to be the spurious result of one sample
of sinaensis (DMNH 67057) lacking
data from two of the three nuclear genes. When analyses are restricted to the
nuclear gene that was sequenced for DMNH 67057 (MUSK), the two samples of sinaensis
form a clade sister to cochabambae, exactly as in the mitochondrial and
combined data (mt + nuclear) trees. See page 14 of the molecular paper.”
Comments from Pacheco: “YES. Recognizing all 16 at the
species level.”
Comments
from Bonaccorso: “NO. It is not that I am not impressed with the huge amount of
relevant data presented by the authors or that I disagree with the core of the
proposal. My problem is that having to consider all these new species together
could be forcing us to make some decisions that we will not make if we were
discussing these splits one by one (or at least a few at the time). I agree
with Santiago that low sample sizes for some species and similarity of songs
for others are problematic, at least with the data at hand.
“I know
this proposal has already passed, but I am asking you to re-consider splitting
this proposal into sub-proposals (one by clade) and vote on them separately, or
at least do so for the clades where species limits are not clear-cut. If
proposed species don´t make the “cut” we can assign one species name to that
clade, based on taxonomic priority, until more evidence is provided to elevate
the subclades to species. I am just trying to save us the trouble of having to
reverse granting species status to some of these entities.”