Proposal (889) to South
American Classification Committee
Change linear sequence
in Scytalopus to reflect phylogenetic relationships
The
present linear sequence of Scytalopus species is originally based on
morphology. The many newly described forms have been thrown in as fit.
Cadena
et al. (2020) presented phylogenetic trees that included all known
species. Additionally, the trees include numerous populations, many of which are
genetically differentiated. In order to avoid future shuffling, this
differentiation was considered in the suggested linear sequence.
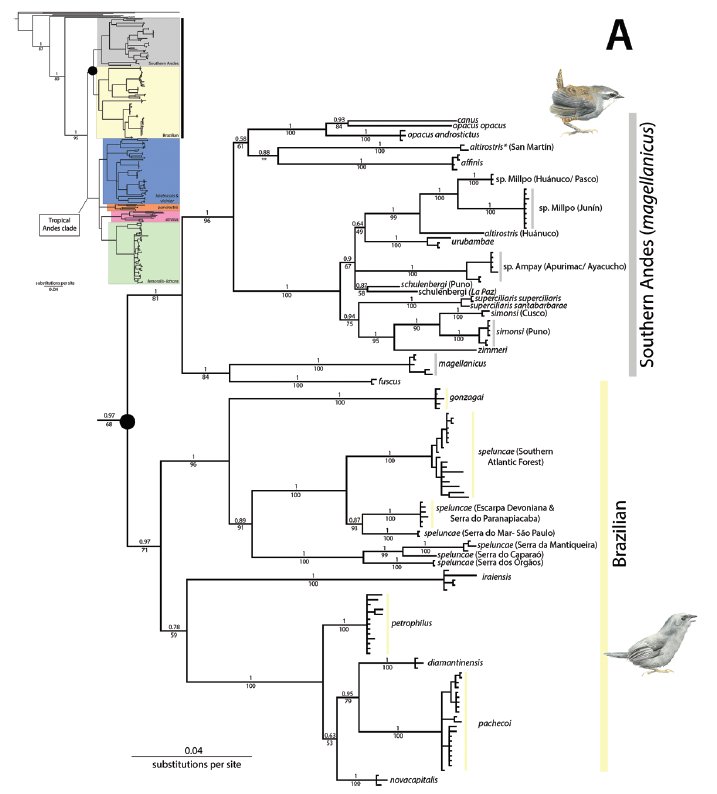
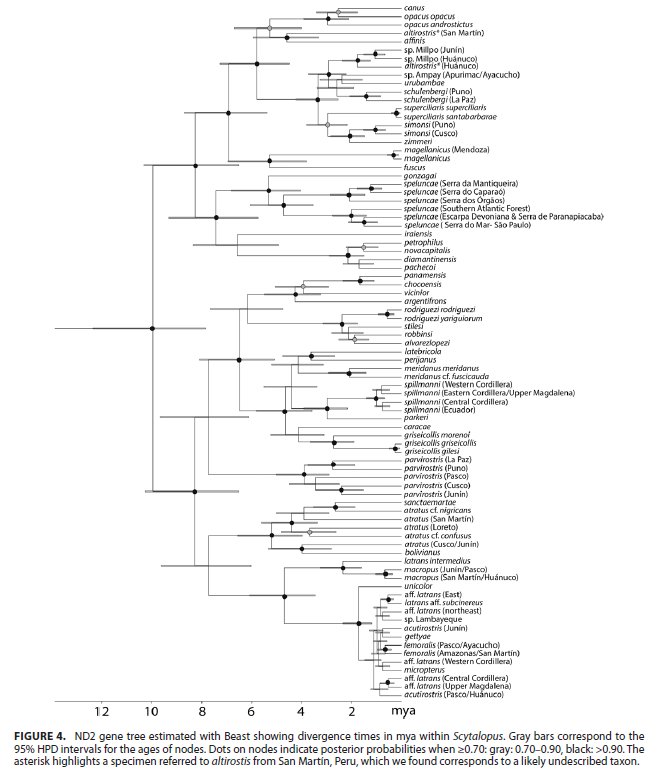
Bayesian
tree showing phylogenetic relationships of Scytalopus tapaculos and allies
inferred using MrBayes based on sequences of the ND2 mitochondrial gene. From
Cadena et al. 2020.
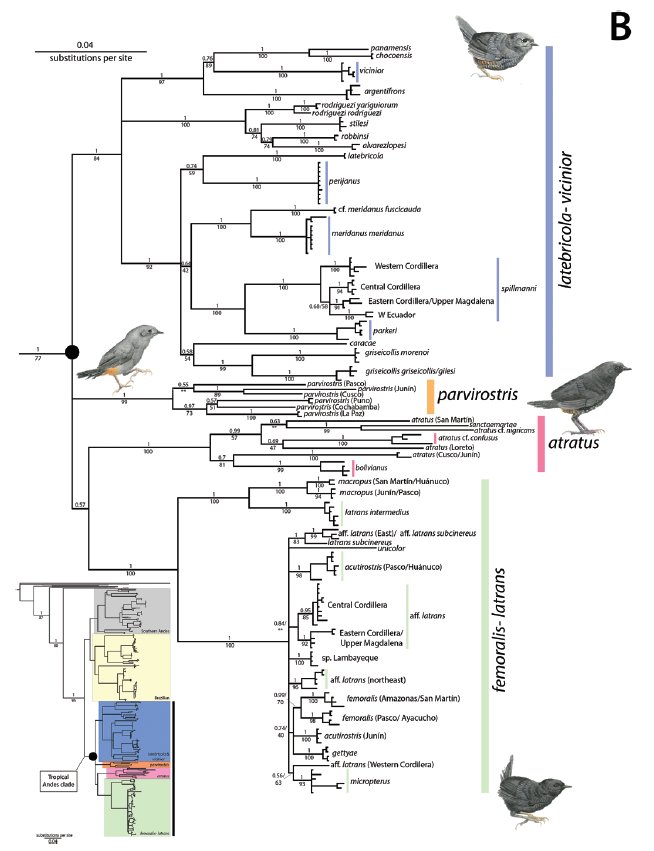
From Cadena et al.
2020.
Notes:
Brazilian
clade: because speluncae shows multiple deep divergences and may
comprise 6 or 7 species (Pulido-Santacruz et al 2016), gonzagai
and speluncae are, with some hesitation, placed after the other
Brazilian branch, which contains 5 species.
Southern
Andes clade: the relationships between schulenbergi, urubambae,
and whitneyi are unresolved. There is strong support for a close
relationship between altirostris and frankeae, which then should
be placed last. Using the simple hypothesis, that geographically closest
allopatric forms within a clade are each other's closest relatives, all five
are then best placed in a south-north sequence. The same justifies placing canus
after opacus.
Tropical
Andes clade: has 4 well-, or fairly well-supported branches: [parvirostris]
(1-6 spp), [atratus] (3-7 spp), [femoralis] (8-16 spp), and [griseicollis]
(16-18 spp).
In
the [atratus] branch Cuzco birds belong with bolivianus, so bolivianus
should come first in the sequence, and it then again makes most sense to list
the species in a south-northward direction, placing atratus between bolivianus
and sanctaemartae.
In
the [femoralis-latrans] branch intermedius and macropus
must come first, followed by unicolor, which appears to be basal to the
rest (see Beast divergence time tree), in which speciation is so recent that
relationships are difficult to assess with certainty. As a general rule,
however, close relatives in Scytalopus tend to have similar elevational
ranges (Cadena & Céspedes 2020), so the high elevation gettyae, acutirostris
and latrans are listed together and right after the high-elevation unicolor.
Latrans may encompass several species and acutirostris two or
three, so gettyae is placed first.
The
[griseicollis] branch has three groups: [argentifrons] (3-4 spp),
[robbinsi] (4-5 spp), and [griseicollis] (7-10 spp).
The
[griseicollis] group has three branches: [griseicollis] (2-3
spp), [latebricola] (3-4 spp) and [spillmanni] (2-5 spp). As griseicollis
may be two species, caracae is placed before it. As spillmanni
may be more than one species, it is placed after parkeri.
The
suggested linear sequence thus becomes:
iraiensis
petrophilus
novacapitalis
diamantinensis
pachecoi
gonzagai
speluncae
fuscus
magellanicus
affinis
krabbei
androstictus
opacus
canus
superciliaris
zimmeri
simonsi
schulenbergi
urubambae
whitneyi
frankeae
altirostris
parvirostris
bolivianus
atratus
sanctaemartae
intermedius
macropus
unicolor
gettyae
acutirostris
latrans
micropterus
femoralis
argentifrons
vicinior
panamensis
chocoensis
rodriguezi
stilesi
alvarezlopezi
robbinsi
caracae
griseicollis
latebricola
perijanus
meridanus
parkeri
spillmanni
References:
Cadena, C. D. and
Céspedes, L. N. 2020. Origin of elevational replacements in a clade of nearly
flightless birds: most diversity in tropical mountains accumulates via
secondary contact following allopatric speciation. Chapter 23 (pp. 635-659) in
V. Rull & A. C. Carnaval, Eds., Neotropical Diversification: Patterns and
Processes. Springer Nature, Basel, Switzerland.
Cadena, C. D., Cuervo,
A. M., Céspedes, L. N., Bravo, G. A., Krabbe, N., Schulenberg, T. S.,
Derryberry, G. E., Silveira, L. F., Derryberry, E. P., Brumfield, R. T., and
Fjeldså, J. 2020. Systematics, biogeography, and diversification of Scytalopus
tapaculos (Rhinocryptidae), an enigmatic radiation of Neotropical montane
birds. The Auk, Ornithological Advances 137: 1-30.
Pulido-Santacruz, P.,
Bornschein, M. R., Belmonte-Lopes, R., and Bonatto, S. L. 2016. Multiple
evolutionary units and demographic stability during the last glacial maximum in
the Scytalopus speluncae complex (Aves: Rhinocryptidae). Molecular
Phylogenetics and Evolution 102: 86-96.
Niels
Kaare Krabbe, October 2020, revised by T. S. Schulenberg
Comments from Areta: “YES. I especially enjoyed the care taken
in considering possible future splits/descriptions to apply the ‘number of
species in a clade’ criterion.”
Comments by Lane: “YES. I think Niels has taken careful consideration
here in this new organization, and I think it has legs.”
Comments by Claramunt: “[NO] The UCE data is the same,
but the proposal didn’t consider much the UCE tree, at least for the Brazilian
clade. In particular, the sequence seems to be based on the Bayesian ND2 tree
but even the ML ND2 tree shows different topology. Some of the nodes are not
well supported in both analyses. I would prefer the following sequence because
it will be compatible with the UCE topology and would also follow the
geographic rule:
S. diamantinensis
S. novacapitalis
S. petrophilus
S. pachecoi
“Not sure if similar cases exist in the Andean clade.”
Comments by Remsen: “NO, not until the above gets
resolved. As long as we’re going to overhaul
the sequence, let’s make sure it best reflects the phylogeny.”
Comments from Zimmer: “YES. This sequence is carefully reasoned and
appears well supported by the genetic, biogeographic and ecological
(altitudinal) data, and nicely anticipates future splits within each clade.”
Response from Niels Krabbe: “Thanks Santiago, for taking time to look closer at this. As to
changing the sequence to accommodate UCE data, I would not recommend it. I am
no expert, but I do find it suspicious that the UCE data differ so much from
the mt data except in the most basal branches. I would
prefer to stick to a mt tree and not consider the UCE data at all for species
level taxonomy. Bayesian mt trees have been used for many of the sequences of
species in SACC. The Bayesian tree gives a 95% posterior probability for a sister
relationship between diamantinensis and pachecoi, so they should
be next to each other in the sequence, unlike what you suggest. That novacapitalis
is sister to them is less supported (65%), so there is reasonable doubt as to
whether novacapitalis should come before or after petrophilus. I
am fine with changing the sequence of these two to follow the NW-SE geography
guideline and give the sequence for this Brazilian branch as:
novacapitalis
petrophilus
diamantinensis
pachecoi
“As to the
different topologies in Bayesian and Maximum Likelihood mt trees, they are
minor. I only refer to the ML tree in one case (the basal position of
unicolor), but probably should not have. The sequence would have come out the
same, so I am happy with striking that reference and base the sequence entirely
upon the Bayesian tree.”
Comments from Bonaccorso: “NO. Although, Niels is right. We
do adjust sequences according to mt DNA all the time. However, in this case,
more data implies conflicts between datasets (something that we don´t have in
many cases). A change in sequence will not have a real impact on most
ornithological studies; why not wait until we get it right?”
New response from Niels Krabbe: “Having taken a closer look at the nuclear data, I realize the strength of it. It would indeed be wrong to disregard it. The nuclear Species Tree Analysis (STA) proves to be a bit of a challenge, as it is at odds with the other three trees in several respects, but there is general agreement between the UCE ML tree and the mt trees. Only three changes to the originally proposed sequence are required: the order of species in the Brazilian clade, and the placements of the micropterus-femoralis branch and gettyae.
“Brazilian clade: All four trees place speluncae and gonzagai together and novacapitalis, petrophilus, diamantinensis and pachecoi together. Both nuclear trees place iraiensis as basal to them all with >90% support, whereas both mt trees place it with poor support as basal to novacapitalis, petrophilus, diamantinensis and pachecoi. There is thus greatest support for placing iraiensis first in the linear sequence as already proposed. A closer look at the mt data indeed reveals great uncertainty as to the relationships of novacapitalis, petrophilus, diamantinensis and pachecoi. Using iraiensis and fuscus as outgroups, 1-6 of 929 bp substitutions tie them together in various combinations: d-n-pe 6, d-pa 5, pa-n-pe 5, d-pa-n 4, n-pe 3, d-pa-pe 1, pa-n 1, and d-n 1. The UCE Species Tree Analysis (STA) also indicates much uncertainty as to these relationships, placing two samples each of novacapitalis and petrophilus in different branches with >90% support, whereas the UCE Maximum Likelihood tree places each of them with the other sample of its own species, also with >90% support. Santiago’s suggestion to place all four in NW-SE direction (of their centers of distribution) seems to be the safest approach, resulting in the linear sequence for the Brazilian taxa being iraiensis, diamantinensis, novacapitalis, petrophilus, pachecoi, gonzagai, speluncae.
“South Andes clade: All four trees agree that fuscus and magellanicus are sister species and basal in the clade. The UCE STA placement of zimmeri with >90% support as being basal to the rest of the clade is at odds with the other three trees. Considering the almost identical plumages, geographical proximity, and fairly similar vocalizations of zimmeri and superciliaris, it is preferable that the suggested linear sequence follows the other three trees (i.e., is maintained as proposed).
“Tropical Andes clade: There is general congruence in all four trees, also in grouping micropterus-femoralis together and close to the unicolor group, but both nuclear trees place them as sisters to that group with strong support, whereas both mt trees place them within it on the basis of much fewer data. They do stand out like a sore thumb from the rest in elevational distribution and body size, so to follow the nuclear trees is a most welcome move.
“One major difference between nuclear and mt trees in the Tropical Andes clade is the placement of gettyae. Both nuclear trees place it basally in the intermedius-macropus clade, whereas the mt trees place it with the rest of the unicolor group. While the ND2 gene shows a very close relationship of macropus with intermedius (41 mostly unique mutations out of 929 bp tie the two together) the mt evidence for a relationship of gettyae with these two forms is rather weak (3 unique mutations of 1041 bp, one of them only to macropus). The nuclear data unequivocally tie gettyae to intermedius and macropus, in fact so strongly, that it highly surprising that it does not show more in the mt data. Whether gettyae is closest to macropus, as weakly suggested by both the UCEs and the mt data, or basal to them, as the sparsity of ties in mt data would suggest, remains unresolved. I thus suggest all three are best placed in a north-south sequence.
“With these three changes the proposed sequence then becomes:
iraiensis
diamantinensis
novacapitalis
petrophilus
pachecoi
gonzagai
speluncae
fuscus
magellanicus
affinis
krabbei
androstictus
opacus
canus
superciliaris
zimmeri
simonsi
schulenbergi
urubambae
whitneyi
frankeae
altirostris
parvirostris
bolivianus
atratus
sanctaemartae
micropterus
femoralis
intermedius
macropus
gettyae
unicolor
acutirostris
latrans
argentifrons
vicinior
panamensis
chocoensis
rodriguezi
stilesi
alvarezlopezi
robbinsi
caracae
griseicollis
latebricola
perijanus
meridanus
parkeri
spillmanni
“I am not sure if these changes require new votes from all reviewers (or even an entirely new proposal) or just acceptance from Van, Elisa and Santiago. In any case, I expect this sequence to reach consensus.”
Comments
from Stiles:
“YES. What Niels decides on this is fine with me! so
YES.
Comments
from Stiles:
“The doubts and suggestions regarding the original proposal for the sequence
were considered in detail and (to the extent that data permit) resolved in a
revised sequence by Niels. Perhaps ask for a re-vote on the revised sequence
(which in any case could be revised in future, but appears to best represent
the current state of knowledge in this most dynamic genus). If NO wins a
majority here, the only option would be to stay with the version in the current
checklist, which is clearly the least satisfactory of the three.”
Comments
from Pacheco:
“YES, for the new sequence proposed by Niels after
the most accurate examination of the nuclear data.”
Additional
comments from Remsen:
“YES. Minor repairs completed.”